Real-Time PCR:
the TaqMan®
Method
This web page was produced as an assignment
for an undergraduate course at Davidson College.
Introduction:
The advent of Polymerase Chain Reaction (PCR)
by Kary B. Mullis in the mid-1980s revolutionized molecular biology as we
know it. PCR is a fairly standard procedure now, and its use is extremely
wide-ranging. At its most basic application, PCR can amplify a small amount
of template DNA (or RNA) into large quantities in a few hours. This is performed
by mixing the DNA with primers on either side of the DNA (forward and reverse),
Taq polymerase (of the species Thermus aquaticus, a thermophile
whose polymerase is able to withstand extremely high temperatures), free nucleotides
(dNTPs for DNA, NTPs for RNA), and buffer. This movie
shows PCR in action. The temperature is then alternated between hot and cold
to denature and reanneal the DNA, with the polymerase adding new complementary
strands each time. In addition to the basic use of PCR, specially designed
primers can be made to ligate two different pieces of DNA together or add
a restriction site, in addition to many other creative uses. Clearly, PCR
is a procedure that is an integral addition to the molecular biologist’s
toolbox, and the method has been continually improved upon over the years.
(Purves, et al. 2001)
Fairly recently, a new method of PCR quantification
has been invented. This is called “real-time PCR” because it allows
the scientist to actually view the increase in the amount of DNA as it is
amplified. Several different types of real-time PCR are being marketed to
the scientific community at this time, each with their advantages. This web
site will explore one of these types, TaqMan® real-time PCR, as well as
give an overview of the other two types of real-time PCR, molecular beacon
and SYBR® Green. (www.ambion.com
2003)
How TaqMan®
works:
TaqMan® utilizes a system that is fairly
easy to grasp conceptually. First, we must take a look at the TaqMan®
probe.
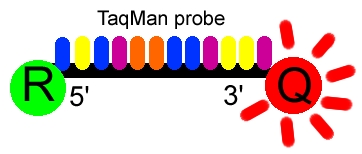
Figure 1. The Taqman probe. The red circle represents the
quenching dye that disrupts the observable signal from the reporter dye (green
circle) when it is within a short distance. Image created by Dan Pierce.
The probe consists of two types of fluorophores,
which are the fluorescent parts of reporter proteins (Green Fluorescent Protein
(GFP) has an often-used fluorophore). While the probe is attached or unattached
to the template DNA and before the polymerase acts, the quencher (Q)
fluorophore (usually a long-wavelength colored dye, such as red) reduces the
fluorescence from the reporter (R) fluorophore (usually a
short-wavelength colored dye, such as green). It does this by the use of Fluorescence
(or Förster) Resonance Energy Transfer (FRET), which is the inhibition
of one dye caused by another without emission of a proton. The reporter dye
is found on the 5’ end of the probe and the quencher at the 3’
end. (www.probes.com
2003)

Figure 2. The TaqMan® probe binds
to the target DNA, and the primer binds as well. Because the primer is bound,
Taq polymerase can now create a complementary strand. Image created by
Dan Pierce.
Once the TaqMan® probe has bound to its
specific piece of the template DNA after denaturation (high temperature) and
the reaction cools, the primers anneal to the DNA. Taq polymerase
then adds nucleotides and removes the Taqman® probe from the template
DNA. This separates the quencher from the reporter, and allows the reporter
to give off its emit its energy. This is then quantified using a computer.
The more times the denaturing and annealing takes place, the more opportunities
there are for the Taqman® probe to bind and, in turn, the more emitted
light is detected. (www.probes.com
2003)
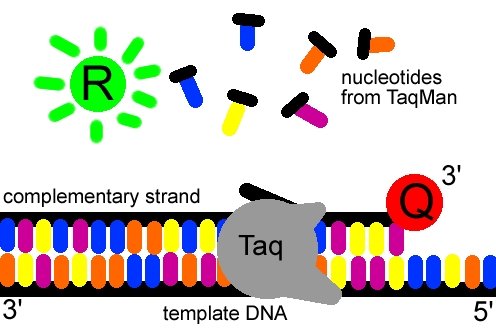
Figure 3. The reporter dye is released from the extending
double-stranded DNA created by the Taq polymerase. Away from the quenching
dye, the light emitted from the reporter dye in an excited state can now be
observed. Image created by Dan Pierce.
Quantification:
The specifics in quantification of the light
emitted during real-time PCR are fairly involved and complex. The math involved
is above the scope of this website, but this website explains some specifics
not talked about here: www.wzw.tum.de.
The light emitted from the dye in the excited state is received
by a computer and shown on a graph display, such as this, showing PCR cycles
on the X-axis and a logarithmic indication of intensity on the Y-axis.
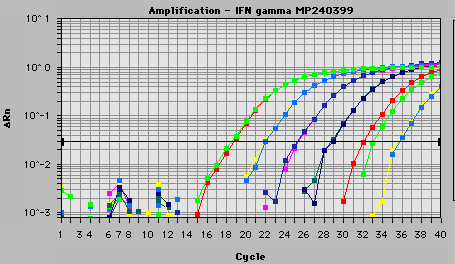
Figure 4. A graph printiout of actual data found using the
TaqMan® probe. Courtesy www.biotech.uiuc.edu.

Figure 5. A real-time PCR machine used at Colorado State.
Courtesy lamar.colostate.edu.
Other Images of TaqMan®
in Action:
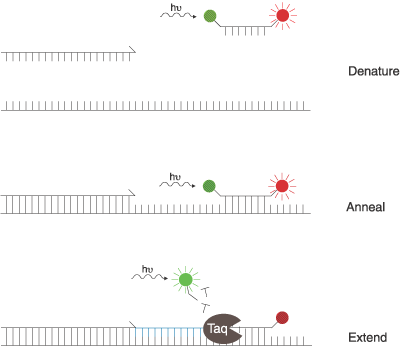
Figure 6. Another three step view of the TaqMan®
probe working: before the probe is met with the Taq polymerase, energy is transferred
from a short-wavelength fluorophore (green) to a long-wavelength fluorophore
(red). When the polymerase adds nucleotides to the template strand, it releases
the short-wavelength fluorophore, making it detectable and the long-wavelength
undetectable. Figure courtesy www.probes.com
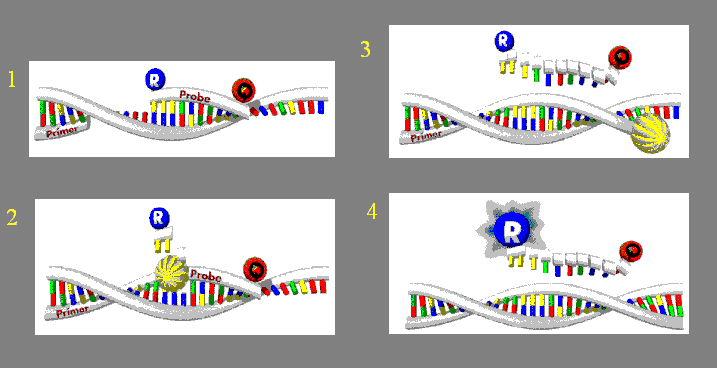
Figure 7. Another view of TaqMan®
in action. The release from the Quencher dye (red Q) in step 2 eventually causes
the Reporter dye (blue R) to be seen in step 4. Figure courtesty www.ruhr-uni-bochum.de
pending.
Other Real-Time PCR Methods:
There are two other types of real-time PCR methods,
the molecular beacon method and the SYBR® Green method. The molecular
beacon method utilizes a reporter probe that is wrapped around into a hairpin.
It also has a quencher dye that must be in close contact to the reporter to
work. An important difference of the molecular beacon method in comparison
to the TaqMan® method is that the probe remains intact throughout the
PCR product, and is rebound to the target at every cycle. Click here to see
a web page on the molecular
beacon method of PCR, another type of real-time PCR used in molecular
biology. The SYBR® Green probe was the first to be used in real-time PCR.
It binds to double-stranded DNA and emits light when excited. Unfortunately,
it binds to any double-stranded DNA which could result in inaccurate data,
especially compared with the specificity found in the other two methods. (www.ambion.com
2003)
References:
Campbell, Malcolm. PCR movie. <http://bio.davidson.edu/misc/movies/pcr2.mov>
Accessed 2/17/03.
Campbell, Malcolm. Real-time PCR, molecular beacon method.. <http://bio.davidson.edu/courses/genomics/method/realtimepcr.html>
Accessed 2/17/03.
Colorado State Laboratory homepage. <http://lamar.colostate.edu/~reddy/labtour2.htm>
Accessed 2/17/03.
Fluorescence Resonance Energy Transfer (FRET). <http://www.probes.com/handbook/boxes/0422.html>
Accessed 2/18/03.
History of PCR. <http://usitweb.shef.ac.uk/~mba97cmh/history/history.htm>
Accessed 2/15/03.
Purves, et al. Life: The Science of Biology. Sixth Edition.
Freeman and Company: USA; 2001. pg. 214-217.
"Real-Time" or Kinetic PCR. <http://dna-9.int-med.uiowa.edu/realtime.htm>
Accessed 2/17/03.
Real-time RT-PCR. <http://www.ambion.com/basics/rtpcr/rtpcr101_page5.html>
Accessed 2/17/03.
W.M. Keck Center for Comparitive and Functional Genomics. <http://www.biotech.uiuc.edu/taqman.htm>
Accessed 2/17/03.
Gene Quantification. <http://www.wzw.tum.de/gene-quantification/relative.html>
Accessed 2/16/03.
Molecular
Biology Main Page
Biology
Home Page
Davidson
College Home Page
Search
Davidson
© Copyright 2003 Department of Biology, Davidson College, Davidson,
NC 28035
Send comments, questions, and suggestions to: dapierce@davidson.edu






