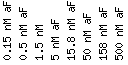This web page was produced as an assignment for an
undergraduate course at Davidson College.
MICROARRAY
ANALYSIS OF
THE YEAST GENE SST2
*Below each data set review the analysis section for an
explanation and discussion of the data presented above*
Experiments:
Expression at different alpha-factor concentrations
Expression in response to alpha-factor
Yeast cell-cycle experiment
Expression during the diauxic shift
Yeast sporulation
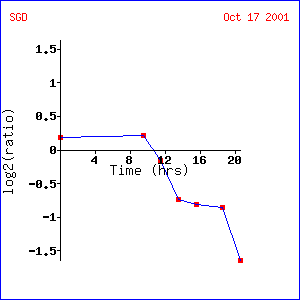
Expression at different alpha-factor concentrations (Rosetta Inpharmatics).
Expression at different alpha-factor concentrations for SST2/YLR452C |
Scale : (fold repression/induction)
![]()
Click on a color strip to see data for that gene.
Up to 20 similar genes are shown, with a Pearson correlation of > 0.8 to the query gene
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
||
|
|
SST2 |
|
|
signal transduction* |
|
GTPase activator |
|
plasma membrane |
||||
|
|
TEC1 |
|
|
pseudohyphal growth |
|
specific RNA polymerase II
transcription factor |
|
nucleus |
||||
|
|
STE2 |
|
|
pheromone response (sensu
Saccharomyces) |
|
mating-type alpha-factor
pheromone receptor |
|
integral plasma membrane
protein |
||||
|
|
FUS3 |
|
|
protein amino acid
phosphorylation* |
|
MAP kinase |
|
nucleus* |
||||
|
|
AGA1 |
|
|
agglutination |
|
cell adhesion receptor |
|
cell wall |
||||
|
|
CIT1 |
|
|
|
tricarboxylic acid cycle* |
|
citrate (SI)-synthase |
|
mitochondrial matrix |
|||
|
|
ECM2 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
||||
|
|
ARG4 |
|
|
arginine biosynthesis |
|
argininosuccinate lyase |
|
cytosol |
||||
|
|
AXL1 |
|
|
axial budding* |
|
metalloendopeptidase |
|
intracellular |
||||
|
|
CNA1 |
|
|
cell wall organization and
biogenesis* |
|
calcium-dependent protein
serine/threonine phosphatase |
|
cytoplasm |
||||
|
|
FUS1 |
|
|
conjugation (sensu Saccharomyces) |
|
molecular_function unknown |
|
plasma membrane |
||||
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||||
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||||
|
|
|
|
|
|
|
|
|
|
||||
|
|
MTC2 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||||
|
|
|
|
|
|
|
|
|
|
||||
|
|
|
|
|
|
|
|
|
|
||||
|
|
PRM1 |
|
|
cell-cell fusion |
|
molecular_function unknown |
|
integral membrane protein* |
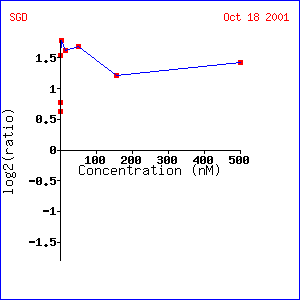
Analysis: SST2 increased gene expression as pheromone concentration increased. Because project 2 already determined SST2’s role in pheromone response (GTPase activator), these results are expected. STE2, a gene involved in pheromone response, is expressed similarly to SST2, which is not surprising because SST2 signals the presence of pheromones by initiating the G-protein response. Other genes, such as AXL1 and FUS1, are logically similar because they are associated with cell reproduction, which is the subsequent response to mating (which is , of course, initiated through pheromones.) TEC1 might be interesting to investigate further to determine whether the pseudohyphal growth response is also involved in reproduction.
* : indicates that more than one annotation exists for the gene.
|
Expression at
different alpha-factor concentrations for SST2/YLR452C |
|
|
![[X]](./SST2draft_files/image048.gif) Expression
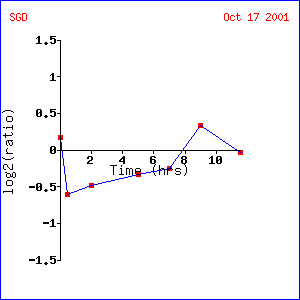
in response to alpha-factor (Rosetta Inpharmatics).
Expression
in response to alpha-factor (Rosetta Inpharmatics).
Expression in response to alpha-factor for SST2/YLR452C |
Scale : (fold repression/induction)
![]()
Click on a color strip to see data for that gene.
Up to 20 similar genes are shown, with a Pearson correlation of > 0.8 to the query gene
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
SST2 |
|
|
signal transduction* |
|
GTPase activator |
|
plasma membrane |
||
|
|
CHS7 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
FUS1 |
|
|
conjugation (sensu Saccharomyces) |
|
molecular_function unknown |
|
plasma membrane |
||
|
|
KAR4 |
|
|
meiosis* |
|
transcription factor |
|
nucleus |
||
|
|
GPA1 |
|
|
signal transduction of mating signal (sensu Saccharomyces) |
|
heterotrimeric G-protein GTPase, alpha subunit |
|
plasma membrane* |
||
|
|
FUS3 |
|
|
protein amino acid phosphorylation* |
|
MAP kinase |
|
nucleus* |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
PRM1 |
|
|
cell-cell fusion |
|
molecular_function unknown |
|
integral membrane protein* |
||
|
|
AGA1 |
|
|
agglutination |
|
cell adhesion receptor |
|
cell wall |
||
|
|
PRM6 |
|
|
mating (sensu Saccharomyces) |
|
molecular_function unknown |
|
integral membrane protein |
||
|
|
PRM4 |
|
|
mating (sensu Saccharomyces) |
|
molecular_function unknown |
|
integral membrane protein |
||
|
|
FAR1 |
|
|
cell cycle arrest |
|
cyclin-dependent protein kinase inhibitor |
|
nucleus* |
||
|
|
STE12 |
|
|
pseudohyphal growth* |
|
transcription factor |
|
nucleus |
||
|
|
HSL7 |
|
|
bud growth* |
|
protein kinase inhibitor |
|
cytokinetic ring (sensu Saccharomyces) |
||
|
|
STE2 |
|
|
pheromone response (sensu Saccharomyces) |
|
mating-type alpha-factor pheromone receptor |
|
integral plasma membrane protein |
||
|
|
SKT5 |
|
|
cytokinesis* |
|
enzyme activator |
|
cytokinetic ring (sensu Saccharomyces) |
||
|
|
FIG2 |
|
|
mating (sensu Saccharomyces) |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SLI15 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
TPD3 |
|
|
protein biosynthesis* |
|
protein phosphatase type 2A |
|
protein phosphatase type 2A |
||
|
|
GIC2 |
|
|
establishment of cell polarity (sensu Saccharomyces)* |
|
small GTPase regulatory/interacting protein |
|
actin cap (sensu Saccharomyces) |
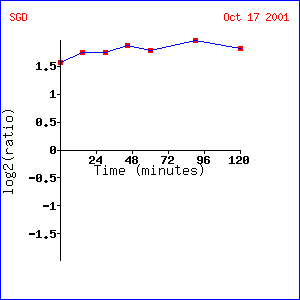
Analysis: The clustering pattern
used in this experiment appears questionable.
Genes CHS7, HSL7, SKT5 and TPD3 do not seem to express similarly to SST2,
which is fully induced at all time points.
Questions are raised about the clustering algorithm used for this
experiment. However, genes that
visually show high induction rates have similar functions to those mentioned above,
namely pheromone response, mating response, cell reproduction. It is interesting to note that the
components of these genes are normally either found in the nucleus or membrane. Cell membrane activity is expected because
the cell is inter-cellularly communicating and must signal through the
membrane. The genes found in the nucleus
seem to relate to transcription or translation which is logical if the genes
they are turning on are also related to pheromone response.
* : indicates that more than one annotation exists for the gene.
|
Expression in
response to alpha-factor for SST2/YLR452C |
|
|
Home | Search | Figures | Data | Info | Links | SGD | Microarray_Homepage
|
Name |
Score |
Peak |
|
|
2.406 |
M/G1 |
|
|
|
3.251 |
M/G1 |
|
|
|
1.71 |
M/G1 |
|
|
|
3.272 |
M/G1 |
|
|
|
0.966 |
|
|
|
|
6.566 |
M/G1 |
|
|
|
3.383 |
M/G1 |
|
|
|
1.342 |
M |
|
|
|
1.3 |
|
|
|
|
3.364 |
M |
|
|
|
3.969 |
G1 |
|
|
|
1.427 |
M/G1 |
|
|
|
|
|
|
|
![[X]](./SST2draft_files/image048.gif) Expression during the diauxic
shift (Stanford University).
Expression during the diauxic
shift (Stanford University).
Expression during the diauxic shift for SST2/YLR452C |
Scale : (fold repression/induction)
![]()
Click on a color strip to see data for that gene.
Up to 20 similar genes are shown, with a Pearson correlation of > 0.8 to the query gene
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
SST2 |
|
|
signal transduction* |
|
GTPase activator |
|
plasma membrane |
||
|
|
GCD1 |
|
|
protein synthesis initiation |
|
translation initiation factor |
|
ribosome |
||
|
|
NUP133 |
|
|
mRNA-nucleus export* |
|
structural protein |
|
nuclear pore |
||
|
|
NAN1 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
FUN12 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
MAD1 |
|
|
mitotic spindle checkpoint |
|
molecular_function unknown |
|
nucleus |
||
|
|
RPF2 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
CYS4 |
|
|
not yet annotated |
|
cystathione beta-synthase |
|
not yet annotated |
||
|
|
ILV6 |
|
|
not yet annotated |
|
acetolactate synthase |
|
not yet annotated |
||
|
|
NSR1 |
|
|
rRNA processing* |
|
RNA binding |
|
nucleolus |
||
|
|
GSP1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
GUA1 |
|
|
not yet annotated |
|
GMP synthase (glutamine hydrolyzing) |
|
not yet annotated |
||
|
|
RNC1 |
|
|
tRNA modification |
|
tRNA methyltransferase |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
FRS2 |
|
|
not yet annotated |
|
phenylalanine--tRNA ligase |
|
not yet annotated |
||
|
|
SCS2 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
RAD6 |
|
|
ubiquitin-dependent protein degradation* |
|
ubiquitin conjugating enzyme |
|
nucleus |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
Analysis: As the diauxic shift increases, the genes clustered here gradually begin to be repressed. Most of the known functions can be placed in three categories: cell growth, signaling or translation. Although this experiment is not exceptionally useful, it is intuitively pleasing because, anthropomorphically, the cell appears to be “prioritizing” and turning off unnecessary genes as food resources become limited.
* : indicates that more than one annotation exists for the gene.
|
Expression during
the diauxic shift for SST2/YLR452C |
|
|
![[X]](./SST2draft_files/image048.gif) Expression during
sporulation (Stanford University).
Expression during
sporulation (Stanford University).
Expression during sporulation for SST2/YLR452C |
Scale : (fold repression/induction)
![]()
Click on a color strip to see data for that gene.
Up to 20 similar genes are shown, with a Pearson correlation of > 0.8 to the query gene
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
SST2 |
|
|
signal transduction* |
|
GTPase activator |
|
plasma membrane |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
TRP4 |
|
|
not yet annotated |
|
anthranilate
phosphoribosyltransferase |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
FLO1 |
|
|
flocculation |
|
not yet annotated |
|
not yet annotated |
||
|
|
ARL1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
AME1 |
|
|
microtubule-based process |
|
molecular_function unknown |
|
spindle pole body |
||
|
|
SRP101 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
FUN11 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
cellular_component unknown |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
cytoplasm |
||
|
|
SPB1 |
|
|
rRNA processing |
|
RNA methyltransferase |
|
nucleus* |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
RPL14B |
|
|
protein biosynthesis |
|
structural protein of
ribosome* |
|
cytosolic large ribosomal
(60S) subunit |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
RPS29A |
|
|
protein biosynthesis |
|
structural protein of
ribosome |
|
cytosolic small ribosomal
(40S) subunit |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
Analysis: SST2 does not show
substantial induction or repression during this experiment as expected. SST2’s known function in pheromone response does
not imply importance in sporulation, therefore, absence of change is not
remarkable.
* :
indicates that more than one annotation exists for the gene.
|
Expression during
sporulation for SST2/YLR452C |
|
|
SGDtm pages Database Copyright © 1997-2001
The Board of Trustees of Leland Stanford Junior University. Permission to use
the information contained in this database was given by the
researchers/institutes who contributed or published the information. Users of
the database are solely responsible for compliance with any copyright
restrictions, including those applying to the author abstracts. Documents from
this server are provided "AS-IS" without any warranty, expressed or
implied.
Search
another gene name | Go to list of Databases
Searched | SGD
Home
Email the author at: amhartman@davidson.edu
Return to my
home page
Return to my
microarray introduction
Return to the Davidson College Biology Department
Return to the Genomics Home Page