This page was created as an assignment for an undergraduate genomics course at Davidson College
.My Favorite Yeast Proteins: Proteomics Techniques
My Favorite Annotated Yeast Gene Tub1 or YML085C:
Summary of Findings from Previous Analyses:
Previous analyses found that the yeast (Saccharomyces cerevisiae) gene Tub1 encodes for alpha-tubulin. Together with beta-tubulin (encoded by Tub2) this protein makes up microtubules, one of the major componants of the cell cytoskeleton (Richards et al., 2000). Microtubules play an especially important role in chromosome movement and are essential during mitosis. Using data from DNA Microarray analysis, I explored expression patterns of Tub1 under a variety of conditions. Tub 1 typically showed expression pattens similar to Tub2 and other microtubule componants (SGD, 2001).
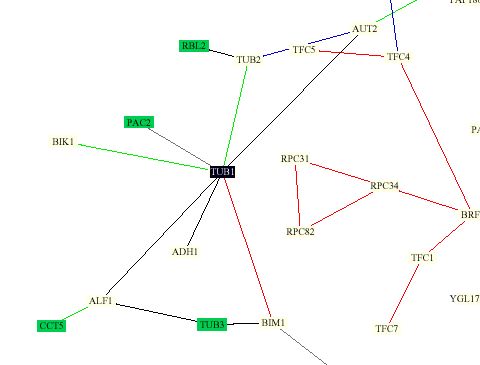
Protein Interaction Map:
This map was constructed by Stan Fields, Peter Uetz and Benno Schwikowski using previously known protein interactions (http://depts.washington.edu/sfields/yplm/data/NB_Figure1color.pdf). The map shows 2,709 interactions between 2,039 proteins. The color scheim for lines is as follows: red = cellular role and subcellular localization of interacting proteins are identical; blue = localizations are identical but functions differ; green = cellular roles are identical but localizations differ; black = cellular role and localization are different or unknown. Proteins in green boxes are involved in cell structure.

It is obvious from the prevelence of green boxes surrounding Tub1 in this figure that Tub1 interacts mainly with proteins involved in cell structure. Not surprisingly, Tub1 interacts with Tub2. One strange aspect of this figure is that they show Tub1 and Tub2 connected with a green line, meaning that they have the same roles indiffering cellular locations. This does not seem correct to me as Tub1 and Tub 2 are expressed simultaneously and for a hertrodiamer(Richards et al., 2000). It would seem that this type of relationship would actually be true for Tub1 and Tub3, as Tub3 is known encode another version of Tub1 (Richards et al., 2000).
In this map Tub1 is also shown to interact with the following proteins. Their functions were obtained from (SGD Function Junction)
Not surprisingly, almost all of the proteins shown to interact with Tub1 are related to microtubule interactions.
Yeast Two-Hybrid Method:
A large-scale yeast two-hybrid screen also identified many other proteins that Tub1 interacts with. This technique attatches half of a transcription factor gene to a "bait" gene. If the protein encoded by the bait gene interacts with a specific "prey" protein, to which the second half of the transcription factor is attatched, the transcription factor initiates the expression of a reporter gene and a subsequent change in color. (http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastList)
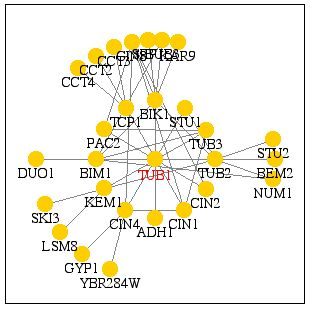
In addition to Tub2, Adh1, Bim1, Pac2, and Bik1 identified above, this method also identified the following proteins that interact with Tub1, functions were obtained from (SGD Function Junction)
Also as expected, the proteins identified using this method were mostly related to the cell cytoskeleton, specifically microtubule-related precesses.
Phospholipid Binding Analysis:
The Gerstein Lab at Yale devoloped a method to detect protein-phospholipid interactions using protein chips. This analysis found that Tub1 did not bind any phospholipids. This makes sense because Tub1 in a structural protein. Interactions with other cellularcomponants (ie phospholipids) are probably regulated through other molecules. (http://spine.mbb.yale.edu/protein_chips/)
Transposon Insertion Method:
The Snyder lab at Yale used random transposon insertion to generate over 11,000 knockout strains of yeast. One of these strains was found to have inserted in the Tub1 gene. They determined from this knockout that Tub1 is an essential gene, as would be expected. They also saw high levels of Tub1 expression during both vegetative growth and sporulation. This would be expected since microtubules are necessary in both of these growth modes. (http://ygac.med.yale.edu/triples/basic_search.asp)
No Useful Information Found At:
DIP database (http://dip.doe-mbi.ucla.edu/dip/Search_DIP.cgi)
PDB database (http://www.rcsb.org/pdb/cgi/explore.cgi?pid=225481005252650&pdbId=1D8B)
My Favorite Unannotated Yeast Gene Lee1 or YPL054W:
Summary of Findings from Previous Analyses:
Lee1 is an unannotated yeast gene whose function has yet to be scientifically investigated. In the previous page BLAST searches discovered two zinc finger regions that lead me to hypothesize that Lee1 may encode a transcription factor. Although DNA microarray data was not terribly conclusive, expression profiles of Lee1 suggest a possible role in DNA replication.
Yeast Two-Hybrid Method:
The yeast two-hybrid assay preformed by Stanley Field's lab found no proteins that interacted with Lee1. This is consistant with Lee1 being a transcription factor as a transcription factor would bind to DNA, not protein. (http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastList)
Phospholipid Binding Analysis:
The phospholipid binding assay preformed by the Gerstein Lab found that Lee1 did not bind any phospholipids. As with the last experiment, this is consistant with Lee1 being a transcription factor. (http://spine.mbb.yale.edu/protein_chips/)
Transposon Insertion Method:
The Snyder lab generated three strains that had transposon insertions in the Lee1 gene. Using these mutants, they foudn that Lee1 was not an esssential gene. Strangely, the three mutants showed conflicting results regarding expression during vegetative groeth and sporultaion. While two strains showed faint or light blue staining (indicating low expression of the LacZ reporter gene contained in the transposon) the third strain showed intense blue staining (indicating high levels of Lee1 expression) under both conditions. I cannot account for these differences, further investigation is necessary. (http://ygac.med.yale.edu/triples/basic_search.asp)
No Useful Information Found At:
Protein Interaction Map (http://depts.washington.edu/sfields/yplm/data/NB_Figure1color.pdf)
DIP database (http://dip.doe-mbi.ucla.edu/dip/Search_DIP.cgi)
PDB database (http://www.rcsb.org/pdb/cgi/explore.cgi?pid=225481005252650&pdbId=1D8B)
Future Experiment 1:
The analyses preformed above were not able to identify any proteins that bound to Lee1. To determine whether or not Lee1 binds to any other proteins I would preform an analysis similar to those done by Gavin MacBeth and Stuart Schreiber at Harvard University (2000). To do this I would attach every protein in the yeast genome to a slide, creating a protein chip of the whole yeast genome. I would then allow Lee1 protein to bind all over the slide, wash, and probe with a flourescent Lee1 antibody. Macbeth and Stuart (2000) have shown that this method is very specific in showing protein-protein interactions. If my predictions are correct, I would not expect Lee1 to bind to other proteins, as it is a transcription factor. It may be though, that this method could identify cofactors that bind with Lee1.
Future Experiment 2:
I would be useful to know the three-dimentional structure of Lee1. It would seem that since Lee1 is a fairly small protein (309 amino acids)(SGD 2001) and does not seem to be membrane-associated (Kyte/Doolittle analysis) it could probably be purified and crystalized. Then its structure could be determined by x-ray crystalography. The structure could then be used to determine binding properties and maybe be used to unravel the interactions of Lee1 with DNA.
Works Cited:
MacBeth, Gavin and Stuart L. Schreiber. (2000). Printing proteins as microarrays for high-throughput function determination. Science 289: 1760-1763.
Richards K.L., et al. (2000) Structure-function relationships in yeast tubulins. Mol Biol Cell. 11:1887-903.
Web Resources:
"Function Junction." (2001) Saccaromyces Genome Database. (http://genome-www.stanford.edu/cgi-bin/SGD/functionJunction)
SGD, Expression connection. (2001) (http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
PathCalling Yeast Interaction Database. (2001) (http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastList)
Gerstein Lab at Yale. (2001) "Global Analysis of Protein Activities Using Proteome Chips" (http://spine.mbb.yale.edu/protein_chips/)
Triples Database (http://ygac.med.yale.edu/triples/basic_search.asp)
DIP database (http://dip.doe-mbi.ucla.edu/dip/Search_DIP.cgi)
PDB database (http://www.rcsb.org/pdb/cgi/explore.cgi?pid=225481005252650&pdbId=1D8B)
Kyte/Doolittle (http://fasta.bioch.virginia.edu/fasta/grease.htm)
SGD (http://genome-www.stanford.edu/cgi-bin/SGD/search)
please email me with questions or comments at jdwillson@davidson.edu