
"This web page was produced as an assignment for an undergraduate course at Davidson College."
Expression of FHL1
The Expression Connection database is part of the Saccharomyces cerevisiae Genome Database (SGD) webservice and it provides access to eleven different DNA microarray experiments. When I typed in the gene name, FHL1, I obtained the following results.
This is the scale used in all of the experiments to show gene expression and repression:

Figure 1. Scale depicting color scheme for gene expression analysis of DNA microarray experiments. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
I performed this search for microarray data on my known gene after I performed my search for my unknown open reading frame (ORF), because I hypothesized that the data for my known gene would be straightforward to analyze since I already knew the molecular function, biological process, and cellular component of the gene. The FHL1 yeast gene encodes a transcription factor which regulates transcription of the RNA Polymerase III promoter, which is involved in rRNA processing. I was wrong. It's not that the data is harder to analyze, but the FHL1 gene doesn't usually cluster with the same genes or other transcription factors and it's not induced or repressed in vary many experiments. It is repressed much more often than it is induced. This is logical because the cell only wants to transcribe the gene encoding this transcription factor when it needs to regulate the RNA Polymerase III promoter, which is involved in rRNA processing. In previous research I learned that a Drosophilia homolog of this transcription factor gene is involved in promoting terminal development. Since this transcription factor is involved in development, it makes sense that it's gene is not induced very often in the following experiments, since none of them focus on factors present in development. Expression levels are slightly altered in various experiments, so it is worth analyzing a few.
This first experiment is observing how expression is regulated by the PHO pathway. Phosphate is an essential nutrient for all cells because it is used to synthesize cellular components like nucleic acids, proteins, lipids, and sugars (DeRisi JL, et al.). The PHO pathway in yeast is a group of regulatory proteins that are activated when extracellular phosphate levels become low. The genes encoding these proteins are induced at these times so that the regulatory proteins can activate the necessary components to obtain more phosphate for the cell. As seen below, FHL1 is repressed in experiment one and in two of the experiments with mutants deficient in accumulating phosphate. This indicates that FHL1 is not important in regulating the PHO pathway in yeast. The FHL1 gene clustered (had a similar expression pattern) with many genes with unknown functions and known genes with a variety of functions. Some of these functions included protein kinases, polymerases, helicases, a structural part of a ribosome, and another transcription factor. Due to the variety of functions of the clustered genes, I don't know that I would be able to hypothesize the function of FHL1 if it was unknown, but as will continue to be obvious, DNA microarray data can provide very valuable information for both genes with unknown and known functions.
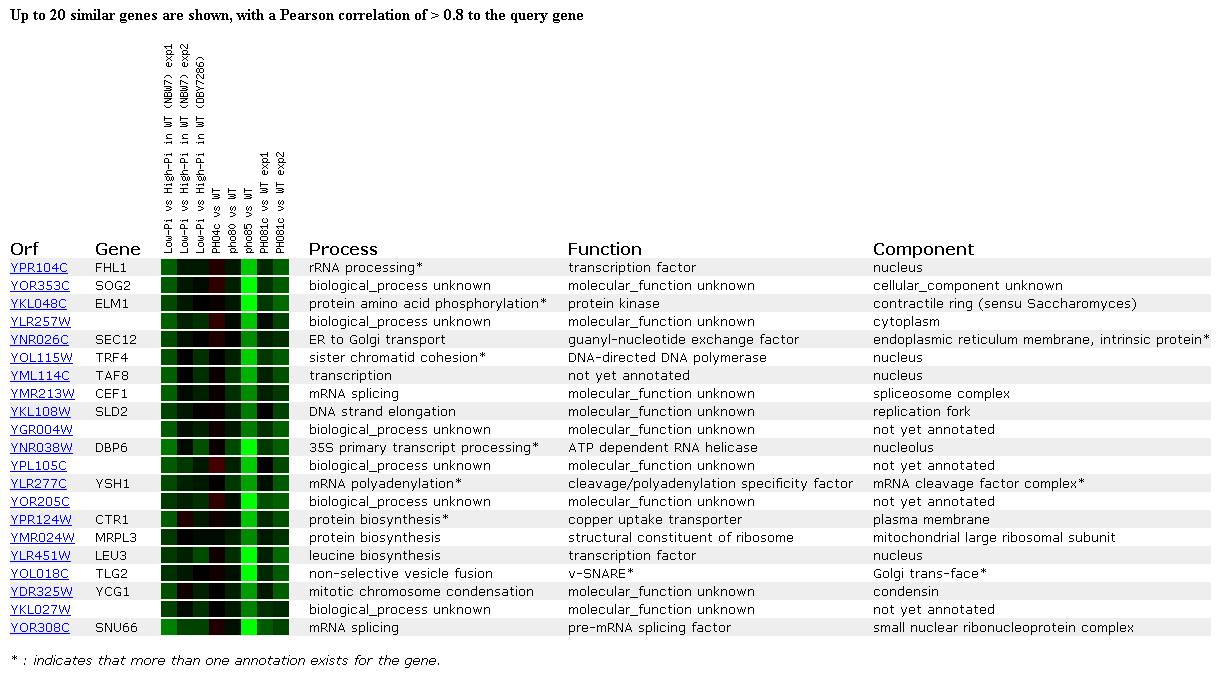
Figure 2. Gene expression regulated by the PHO pathway. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to read the paper associated with the PHO pathway data.
Another useful experiment observed the effects of MANY environmental changes on the yeast genome. These environmental changes include various methods of heat shock, the addition of sorbital, hydrogen peroxide, DTT, diamide, or menadine, hypo-osmotic shock, amino acid starvation, nitrogen depletion, a diauxic shift, stationary phase, gene overexpression, exposure to sugar, and steady state experiments. This is an impressive number of experiments for one group to perform and although it provides a large amount of data, the FHL1 gene was repressed in most of the experiments in which there was any change from normal expression and this gene didn't cluster with any other genes. Expression of FHL1 was repressed in the initial heat shock experiment, particullarly at 15 and 20 minutes and also in another heat shock experiment where the temperature was raised to 37 degrees celcius. Strong repression was also observed when the cells were exposed to low levels of hydrogen peroxide, long periods of exposure to DTT, and during the stationary phase experiments. Less intense repression was also seen with long periods of exposure to menadione and low levels of diamide. A small level of induction is observed after 5 days of nitrogen depletion. I think that the most important part of this DNA microarray data is the repression of FHL1 during low levels of hydrogen peroxide, because I find it interesting that this gene for a transcription factor is repressed initially and then reaches normal levels of expression as a stress, hydrogen peroxide, remains in the cellular environment for longer periods of time. I think that some of the other experiments are also probably interesting, but currently, I don't know enough about the effects of the different chemicals to make many precise conclusions.
Click here to read the paper associated with this DNA microarray experiment on the effect of environmental factors on yeast genes.
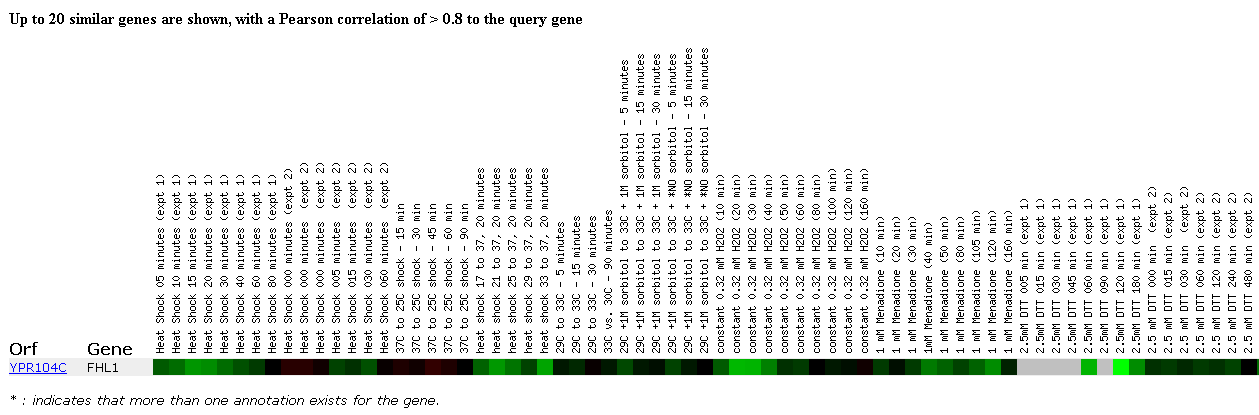
Figure 3. Expression in response to environmental changes. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
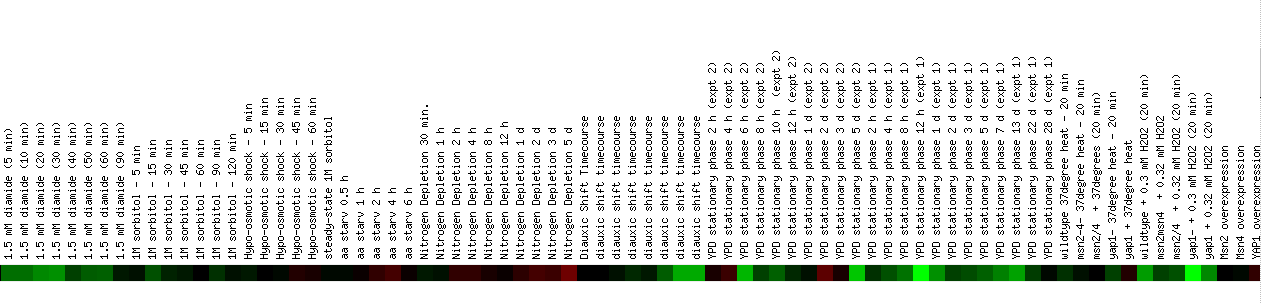
Figure 4. Continued expression of FHL1 in response to environmental changes. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to see the rest of the data. It was too large to fit on the screen.
Expression of FHL1 is also repressed in the following experiment studying gene expression in response to histone depletion. Eukaryotic genomes are packaged around nucleosomes which are believed to repress certain genes (Wyrick JJ, et al.). Histones are also believed to form a complex ('Sir' complex) that silences genes, especially those located on telomeres. Histones and silencing factors were depleted in this experiment, so the researchers hypothesized that most genes would become induced under these conditions. Instead, they found that most genes were not effected at all and 15% were induced and 10% were repressed. Therefore, Wyrick et. al. concluded from this experiment that histones make Sir-independent contributions to telomeric silencing and effect chromosomes in a gene specific manner rather than a generally repressive manner. FHL1 is initially slightly repressed at time zero and when the histone is first depleted and then its expression returns to normal until it is strongly repressed at the final two time points (4 and 6 hours). I conclude from this that the FHL1 gene is not located close to a telomere. Interestingly, 8 out of 20 genes with similar expression patterns in this experiment are structural constituents of ribosomes. I'm not sure what the connection is, but upon depletion of histones, genes encoding ribosomal proteins seem to be expressed in similar patterns.

Figure 5. Expression in response to histone depletion. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to read the abstract of the paper associated with the histone depletion DNA microarray data.
In the next experiment, gene expression was monitored as cells switched from anaerobic to aerobic functions upon depletion of their glucose sources and thus, the cells underwent a metabolic switch from fermentation to respiration. As seen below, FHL1 is repressed after 18.5 hours and 20.5 hours under this aerobic condition. Interestingly again, 10 of the 20 genes with similar expression in this experiment are ribosomal proteins. Upon searching the paper, there were quite a few references involving ribosomes, but I didn't have a chance to read them. Repression of ribosomal proteins appears to be important when yeast switch from anaerobic to aerobic conditions.
Click here to read the full paper on this diauxic shift experiment.
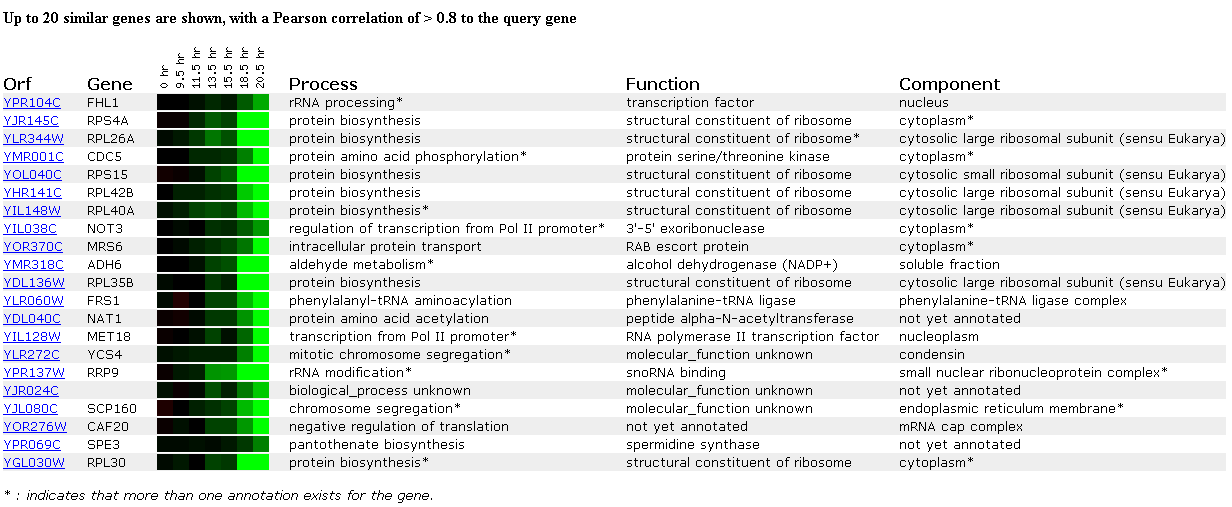
Figure 6. Expression during the diauxic shift. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
In another experiment, gene expression was observed in response to varying zinc levels. Zinc is an essential nutriet for cells and has many functions, but the method of its uptake and the genes affected by its concentration remain unknown. In this experiment, gene expression in wildtype cells, both replete and deficient of zinc, were compared to gene expression in zap1 mutants. Zap1 is a transcription factor which senses zinc dificiency and subsequently increases the expression of certain target genes.
FHL1 expression was slightely induced when wildtype cells were compared to the deficient mutant at 61 nM of zinc and was slightly repressed when the zap1 mutant was exposed to 10 micromoles of zinc. Since not much change in gene expression is observed in this experiment, a conclusion can not be made about the relationship between FHL1 and zinc, but since FHL1 is induced in wild-type cells and repressed in zinc-limited zap1 mutants, there is a small possibility that FHL1 is partially regulated by zap1. A few other ribosomal genes clustered with FHL1 in this experiment, but most of the clustered genes had unknown functions, so not much information was gained from this experiment.
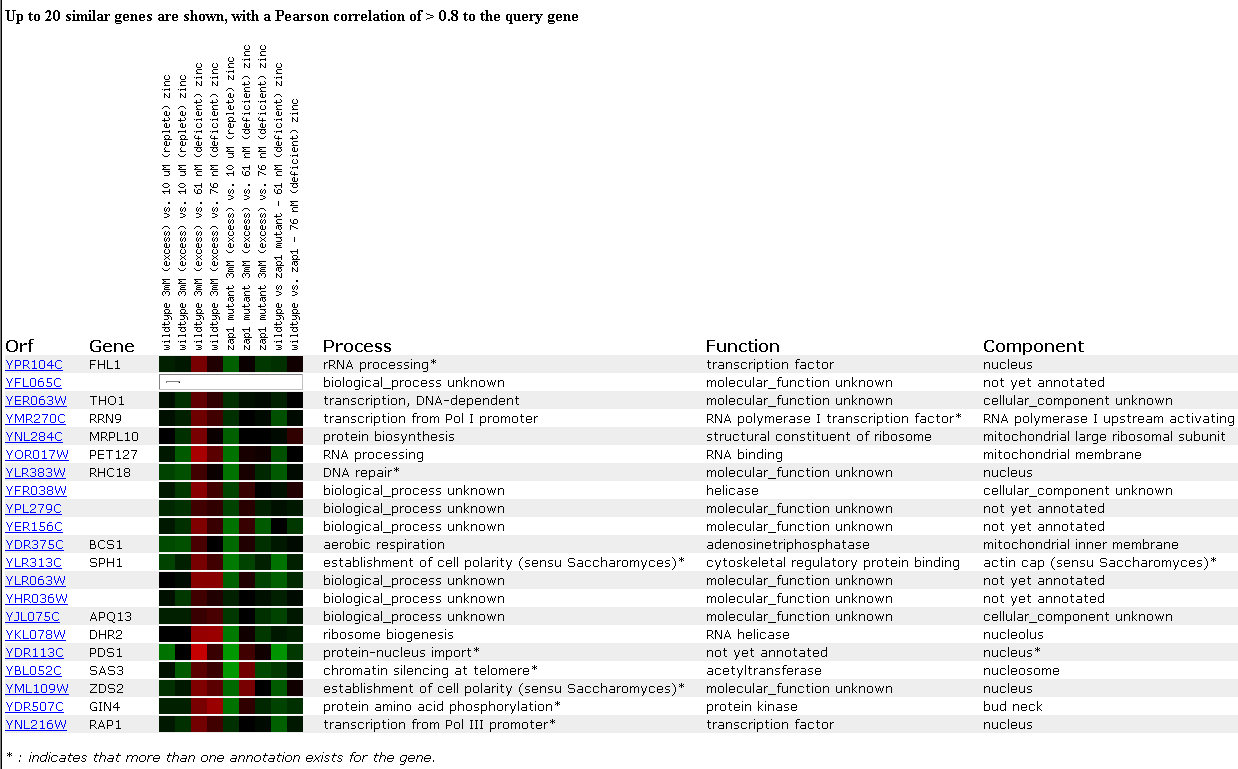
Figure 7. Expression in response to varying zinc levels. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to read the paper published on the effect of zinc levels on the yeast genome.
Although the gene expression of FHL1 is not altered very much in this cell cycle experiment, I wanted to discuss the data because the experiment is set up well and FHL1 gene expression is not changed much in any of the remaining experiments. The goal of the yeast cell cycle analysis project was to identify all genes whose mRNA levels were regulated by the cell cycle (Spellman et al.). Multiple experiments were performed including exposing the cells to cell cycle regulators, exposing temperature mutants to increased temperatures, and arresting alpha-factor exposure over time. FHL1 was slightly induced after 40 minutes of exposure to cell cycle regulators and was repressed after 110 minutes. FHL1 was slightly repressed in the temperature sensitive mutants after being exposure to high temperatures for 200 minutes. Finally, FHL1 was slightly repressed at the beginning time points of the alpha-factor arrest experiment. Transcription of FHL1 might be partially regulated by other transcription factors, but from this experiment, I think it is pretty obvious that FHL1 transcription is not regulated by the cell cycle.
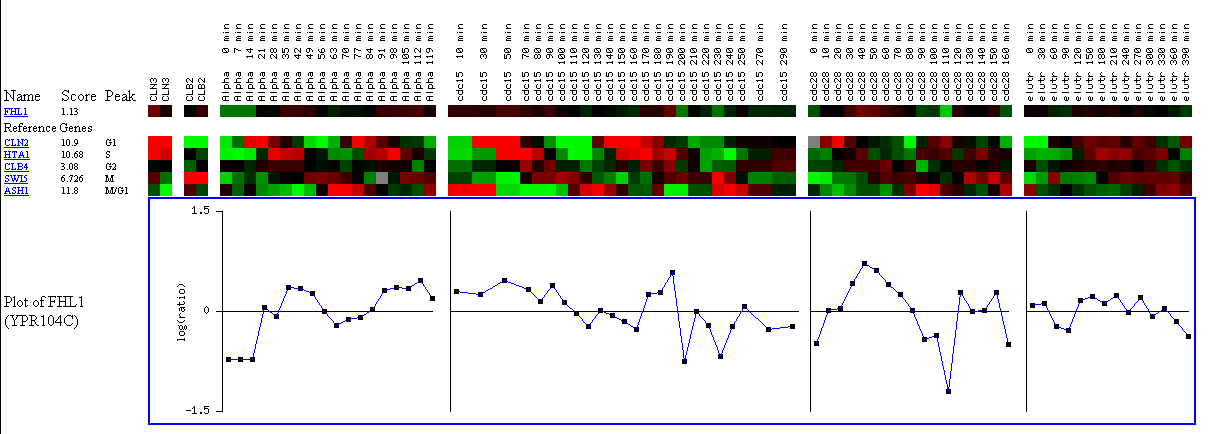
Figure 8. Expression during the cell cycle. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/expression/expressionConnection.pl)
Click here to read the paper published about the yeast cell cycle analysis project.
Additional information can also be obtained at another SGD website named Function Junction. The yeast microarray global viewer section of this webpage provides data from a large number of experiments involving the specific gene of interest entered by the user. This information can be very helpful, but not much information is included about the experimental conditions, so sometimes the results are difficult to interpret.
In the first experiment (Angus-hill--Rsc3-Rsc30), FHL1 is initially repressed and then induced. This experiment involves mutant chromatin remodeling complexes and I would be interested to learn more about the experimental conditions that produced the results shown in figure 8. Click here to read the abstract for this paper.
In glancing at the experiments on the Function Junction webpage, I noticed that FHL1 was repressed in most experiments. The FHL1 gene was repressed in the stress, sporulation, metabolic, Rpn4, CTD, SAGA, and salt experiments. The FHL1 gene was induced in the mitotic and A kinase experiments. As I stated previously, I was initially surprised by these results, but since the FHL1 gene is a transcription factor involved in the regulation of rRNA processing that occurs most often in development, these results are consistent with the known function of the gene.
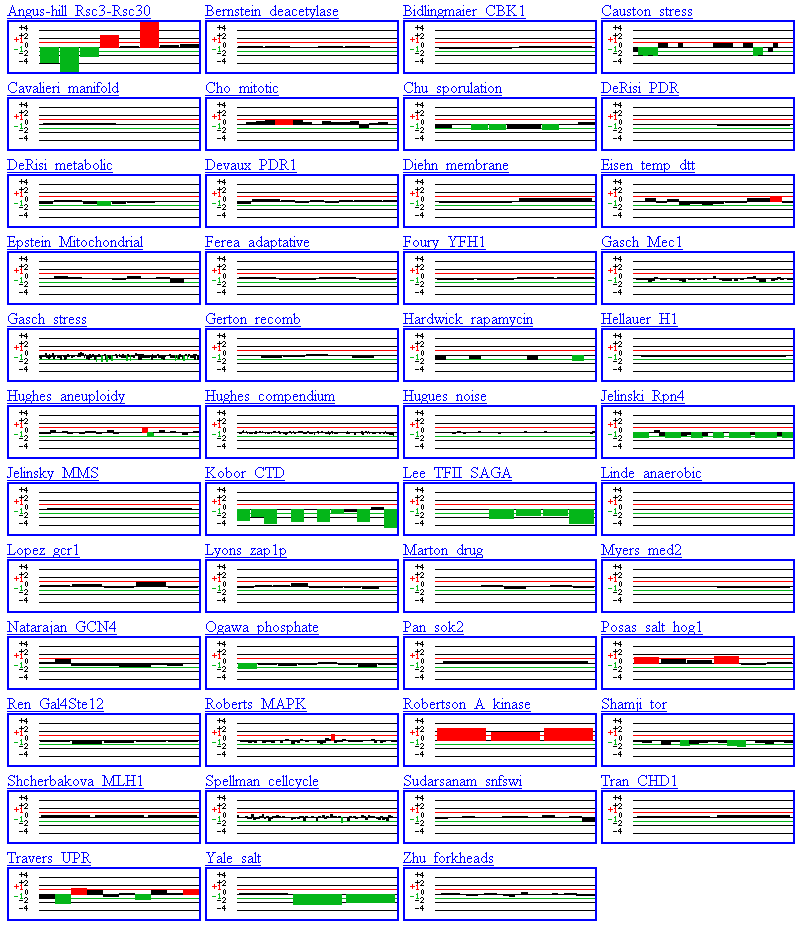
Figure 9. DNA Microarray experiments performed involving the FHL1 gene. Summaries of the results are observed in the small rectangles under the name of the experiment or researcher. (SGD, 2002; http://genome-www4.stanford.edu/cgi-bin/SGD/functionJunction)
Click here to access the Function Junction webpage
Conclusions: Since the molecular function, biological process, and cellular component of the FHL1 yeast gene have already been determined, the purpose of mining DNA microarray databases for information on this gene was to become familiar with the databases and see if any additional information could be found about this gene. Most of the DNA microarray experiments confirmed that the FHL1 gene is most similar to both ribosomal proteins and transcription factors. This is logical since FHL1 encodes a transcription factor involved in rRNA processing. The transcription of the FHL1 gene was repressed in most experiments, which again is logical because the cell doesn't want transcription factors inducing transcription of other genes during normal conditions. Induced gene expression of the FHL1 gene should only be observed when the cell wants to regulate rRNA processing. Induction of the FHL1 gene was observed in mitotic and A Kinase experiments, which again confirms the role of the gene because a transcription factor will probably be activated during mitosis and phosphorylation experiments. Therefore, although I didn't obtain any amazing insights into new roles for the FHL1 yeast gene, the gene's role in the cell was supported by the various DNA microarray experiments.
References:
DeRisi JL, et al. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278(5338):680-6.
Gasch AP, et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11(12):4241-57.
Lyons, TJ, et al. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. PNAS 97 (14): 7957-7962.
Ogawa N, et al. (2000) New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell 11(12):4309-21.
Spellman et al., (1998) Comprehensive Identification of
Cell Cycle-regulated Genes of the Yeast Saccharomyces cerevisiae by Microarray
Hybridization. Molecular Biology of the Cell 9: 3273-3297.
Wyrick JJ, et al. (1999) Chromosomal landscape of nucleosome-dependent
gene expression and silencing in yeast. Nature 402(6760):418-21.
Send comments, questions, and suggestions to: sahenry@davidson.edu