This site was created for an undergraduate Genomics course at Davidson College
Biological Process
The overall objective of yeast gene SSL2 (aka RAD25) is to take part in a process that fixes 'broken' DNA. In particular SSL2 is involved in nucleotide excision repair (NER) (Prakash & Prakash, 2000). There are numerous groups of proteins that work together to perform NER, and these assemblies are called nucleotide excision repair factors (NEF's). Of interest to us is NEF3 which is made up of two sub-assemblies: TFIIH and Rad2 (Habraken et. al., 1996). TFIIH is a general transcription factor made up of 9 proteins, including SSL2 (Myer & Young, 1998), and Rad2 is a nuclease. Habraken. et. al. determined that this association exists because Rad2 is unable to locate UV damaged DNA by itself. They went on to show a tight association between TFIIH and Rad2, and thus concluded that TFIIH is responsible for locating damaged DNA. Prakash & Prakash (2000) point out that there is still a lot to learn about the mechanisms of NER, "the underlying mechanisms by which damage is recognized, NEFs are assembled at the damage site, and the DNA is unwound and incised, remain to be elucidated." I mentioned initially that SSL2's biological process is to fix broken DNA. While there are many ways DNA can become damaged, papers dealing with NER seem to focus on damage caused by UV light and Cisplatin.
Another important aspect of SSL2 is that it is required for yeast to be viable (Giaever et. al. 2002). These researchers performed a huge deletion experiment in which they test most yeast genes to learn whether or not they are required for yeast to be viable. Because of this test we gain some insight into just how important SSL2 is in the TFIIH sub-assembly even though there is another DNA helicase included in this assembly.
Molecular Function
As mentioned above TFIIH is a general transcription factor made up of 9 proteins that perform NER. SSL2 is a DNA Helicase and thus its molecular role in NER is to unwind DNA and form the 'bubble structure' that is necessary for NER. There is another subunit of TFIIH, Rad3, that is also a helicase; however, difference in function between the two DNA helicases is not discussed in the literature. Gene Ontology describes the action of DNA helicase as "The unwinding, or local denaturation, of the DNA duplex to create a bubble around the site of the DNA damage."
Cellular Component
Because SSL2 is a subunit of a transcription factor, the site of its function is the nucleus of the cell. More specifically SSL2 groups together with the TFIIH transcription factor which is localized to damaged DNA by an unknown mechanism.
Restriction Map (Diagram click 'display map')
Chromosomal Map and Combined Physical and Genetic Map
Blastp Results
I performed a Blatp search using the yeast SSL2 protein sequence and recieved a huge number of 'real' hits. It is interesting to note that the results for Yeast SSL2 from Blast showed only 95% positives to the SGD sequence for SSL2. The SGD sequence contains three stretches of amino acids not included in the Blast results SSL2 and Rad25. From this Blast I learned that this gene is highly conserved across species. Included in the list of species with high levels of homology are Drosophilia, Mice, Humans, Schizosaccharomyces pombe, Dictyostelium discoideum, Ciona intestinalis, Anopheles gambiae, C. elegans and many others.
Structural Information
There are no 3D or 2D images of SSL2 currently available. However, I will include a 3D Chime image of a DNA Helicase that may resemble in structure a yeast DNA helicase.
Figure 1: Structure Of DNA Helicase from Bacillus stearothermophilus.
Image provided by PDB and is available at http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1PJR
Predator Secondary Structrure Prediction

Figure 2: Secondary structure prediction performed by NPS Predator.
Random coil : 472 is 55.99% (Yellow)
Alpha helix : 228 is 27.05% (Blue)
Extended strand : 143 is 16.96% (Red)
Kyte-Doolittle Prediction
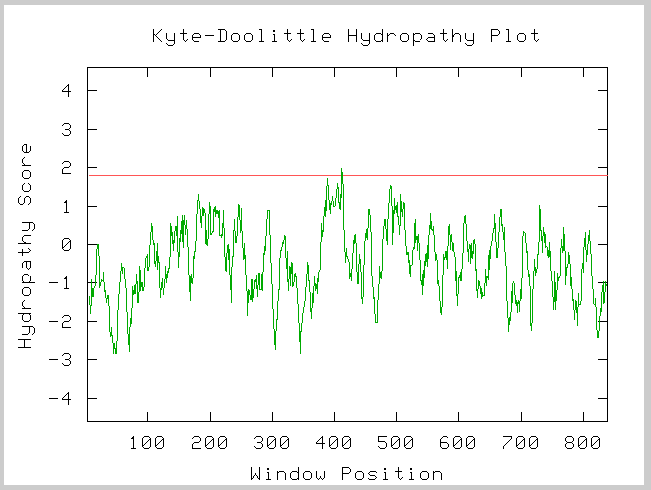
Figure 3: Kyte-Doolittle Hydropathy plot of the amino acid sequence of yeast SSL2.
Image provided by http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm
This Kye-Doolittle hydopathy plot confirms what we know about the localization of SSL2 in yeast. We see one possible transmembrane domain, but it is not very strong. We know that SSL2 is not a membrane bound protein and thus we did not expect to see any transmembrane domains.
Human Genome Browser Search
I searched the human, mouse, and fish genomes using the Human Genome Browser for the human homolog of SSL2, ERCC3. The following figure shows the level of homology that is retained in these three species much as we saw with the Blastp figure above.
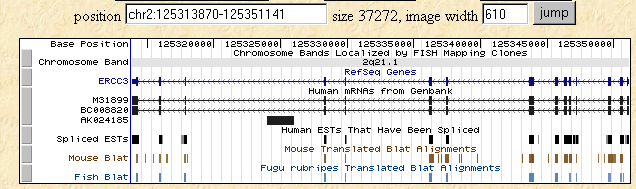
Figure 4: Human Genome Browser map of ERCC3 along with Mouse and Fish Blat data.
This image was taken from http://genome.ucsc.edu/cgi-bin/hgTracks?position=M31899&hgsid=10474682
Another way to view this protein is by looking at its conserved domains. Doing this shows once again the homology between yeast SSL2 and human XPB/ECCR2
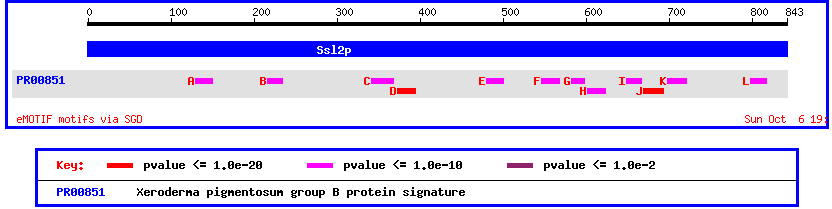
Figure 5: Motif map of yeast SSL2 showing conserved domains with Xeroderma pigmentosum group B protein.
Mutant Alleles of SSL2
A number of papers have explored the affects of mutated forms of SSL2. Work by Ishii et. al. (2000) described the role SSL2 has in maintenence of telomere integrity. In yeast with a mutant form of SSL2 telomeres were gradually shortened in the presence of cisplatin. When they performed a complementation test with wt SSL2 the telomeres retained their integrity. Lee et. al. (1998) found that a yeast with mutant SSL2 allows for increased retrotransposition of Ty1. They hypothesize that SSL2, along with ther NER subassemblies, affect Ty1 postranslationally by affecting reverse transcription or by destabilizing the DNA. Finally, Sweder et. al. (1994) studied an SSL2 mutant where the mutation affected the COOH end of the protein. They concluded that the COOH terminus is required for functioning SSL2.
This is a non-annotated ORF located near SSL2 on yeast chromosome 9.
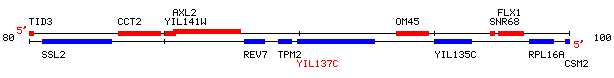
Figure 5: ORF Map of portion of yeast chromosome 9 coordinates 79948 to 102788.
Image available at http://genome-www4.stanford.edu/cgi-bin/SGD/ORFMAP/ORFmap?sgdid=S0001399
One very interesting piece of information that is already availalbe about this ORF is that it is not required for yeast to be viable (Giaever et. al. 2002).
From this homolog information we can learn a lot about the possible function of this unidentified ORF. Using SGD's mammalian homology page we see that YLI137C is homologous to an aminopeptidase/oxytocinase in mice, rats, and humans. Another important piece of the puzzle is given by the SGD eMOTIF database.
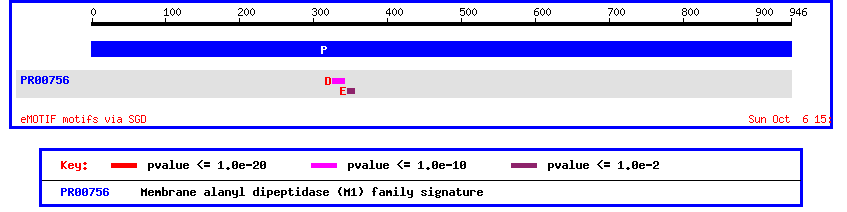
Figure 6: Motif analysis of yeast ORF YIL137C.
Image provided by SGD at http://genome-www4.stanford.edu/cgi-bin/SGD/singlepageformat?sgdid=S0001399#motifs
This motif display shows a membrane alanyle dipeptidase signature. This seems to be in step with the homology information above that predicited this ORF to be some type of aminopeptidase. Since this motif analysis leads us to believe that the unknown ORF is membrane bound, we should check using Kyte-Doolittle.
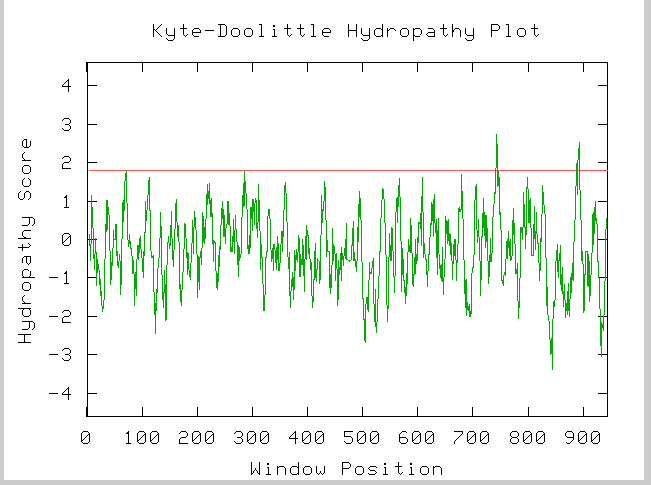
Figure 7: Kyte-Doolittle hydropathy plot for YIL137C amino acid sequence.
Image provided by http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm
From this plot we could hypothesize that YIL137C does in fact have 2 tranmembrane domians at amino acids ~720 and ~890.
Blast2p Analysis
To analyze the degree of similarity between yeast YIL137C and human aminopeptidase I performed a Blast2-protein analysis.
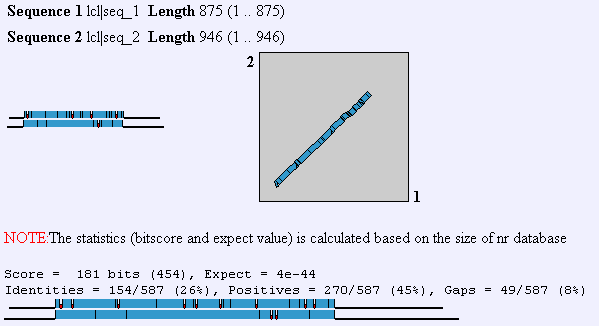
Figure 8: Blast2p comparison of amino acid sequence of yeast YLI137C and human aminopeptidase protein.
Sequence 2 is YLI137C and Sequence 1 is Human aminopeptidase.
This figure was created by NCBI Blast2
We see from this figure a very high level of conservation. I next decided to run a Blast2n for these two proteins. I was very surprised to find that Blast2n found no significant similarity in the two nucleotides sequences whose amino acid sequences are very much homologous. I assume this shows that while the protien structure was maintained over evolutionary time, the bases used to encode the amino acids incurred silent mutations.
I next performed a Blastp with the sequence of YIL137C. It gave me the following motif data:
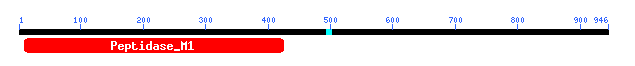
Figure 9: Motif data for yeast ORF YIL137C.
Image provided by NCBI Blast
We learn that this motif is of the Peptidase M1 family which includes aminopeptidases. Check this out at Conserved Domain Database.
Also note the results of my Blastp search which found many real hits for aminopeptidases and oxytocinases.
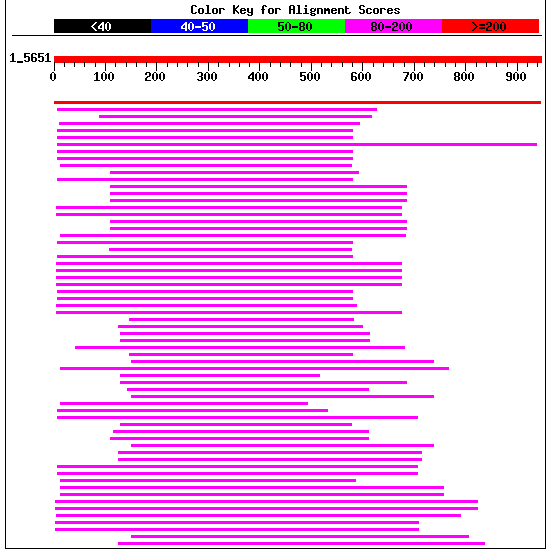
Figure 10: Blastp results for yeast YIL137C.
Image provided by NCBI Blast.
3D Structures of Homologs of YIL137C
We learned from the conserved domain database that leukotriene-A4 hydrolase is a member of the Peptidase M1 family and that the unknown ORF YLI137C contains conserved domains of a leukotriene-A4 hydrolase. I have found a 3D chime file of such a protein.
Figure 11: Structure Of [E271Q] Leukotriene A4 Hydrolase
File provided by PDB at http://www.rcsb.org/pdb/cgi/explore.cgi?job=download&pdbId=1H19&page=
Another usefule image is our unknown ORF YLI137C alligned with the Leukotriene A4 Hydrolase that is in Figure 11.
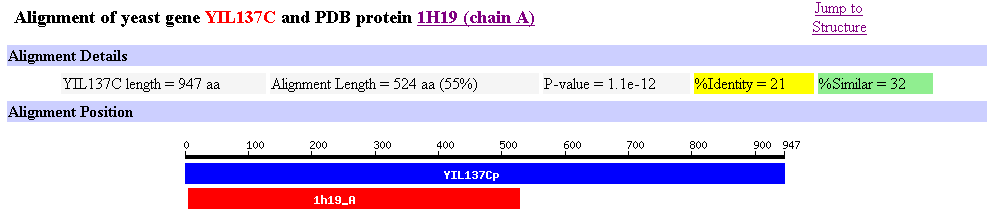
Figure 12: Allignment of yeast ORF YIL137C and Leukotriene A4 Hydrolase.
Image provided by SGD at
http://genome-www4.stanford.edu/cgi-bin/SGD/protein/get3d?locus=YIL137C&pdb=1h19_A&align=1
Summary of Predictions about YIL137C
From the data presented above we can make predictions about the role of YIL137C in yeast. It seems that it is most likely a leukotriene A4 Hydrolase. Kull et. al. (2001) describe the role of a yeast leukotriene A4 hydrolase, but do not mention its cytogenetic location. The clone they describe seems at least related to this unknown ORF. I hypothesize, based on the domain conservation in this ORF, that YIL137C performs the role of an leukotriene A4 hydrolase which is to "catalyzes the hydrolysis of leukotriene A(4) into the proinflammatory mediator leukotriene B(4)" (Kull et. al. 2001), Another paper about these hydrolases is Haeggestrom, 2000.
REFERENCES
Prakash S and Prakash L (2000) Nucleotide excision repair in yeast. Mutat Res 451(1-2):13-24
Giaever G, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418(6896):387-91
Yang WL, Cvijic ME, Ishii K, Chin KV. The requirement of yeast Ssl2 (Rad25) for the repair of cisplatin-damaged DNA. Biochem Biophys Res Commun. 1998 Sep 29;250(3):593-7.
Habraken Y, Sung P, Prakash S, Prakash L. Transcription factor TFIIH and DNA endonuclease Rad2 constitute yeast nucleotide excision repair factor 3: implications for nucleotide excision repair and Cockayne syndrome. Proc Natl Acad Sci U S A 1996 Oct 1;93(20):10718-22
Myer VE and Young RA (1998) RNA polymerase II holoenzymes and subcomplexes. J Biol Chem 273(43):27757-60
Lee BS, Lichtenstein CP, Faiola B, Rinckel LA, Wysock W, Curcio MJ, Garfinkel DJ.Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 1998 Apr;148(4):1743-61
Sweder KS, Hanawalt PC.The COOH terminus of suppressor of stem loop (SSL2/RAD25) in yeast is essential for overall genomic excision repair and transcription-coupled repair. J Biol Chem 1994 Jan 21;269(3):1852-7
Ishii K, Yang WL, Cvijic ME, Kikuchi Y, Nagata I, Chin KV. Telomere shortening by cisplatin in yeast nucleotide excision repair mutant. Exp Cell Res. 2000 Feb 25;255(1):95-101.
Kull F, Ohlson E, Lind B, Haeggstrom JZ. Saccharomyces cerevisiae leukotriene A4 hydrolase: formation of leukotriene B4 and identification of catalytic residues. Biochemistry 2001 Oct 23;40(42):12695-703
Haeggstrom JZ. Structure, function, and regulation of leukotriene A4 hydrolase. Am J Respir Crit Care Med 2000 Feb;161(2 Pt 2):S25-31
Contact Kevin James with any comments or questions.