This web page was produced as an assignment for an undergraduate course at Davidson College.
The Role of Calmodulin in S. cerevisiae
The purpose of this website is to use internet resources to learn more about the proteins encoded by the S. cerevisiae genes CMD1 and YBR108W. CMD1 encodes for calmodulin, which is a calcium binding protein that is fairly active within the cells of numerous organisms. In yeast, some of the processes it participates in are mitosis, endocytosis, and stress response (see CMD1: My Favorite Yeast Gene for more information). YBR108W, on the other hand, is an unannotated ORF. Although microarray data observed in the previous assignment suggested that this gene might be involved in cellular organization somehow, nothing conclusive was revealed.
DIP:
Figure 1: Screen capture of graph generated by DIP showing calmodulin node (centered red dot) and its related protein-protein interactions. The cluster of nodes around calmodulin includes the proteins ILS1, ORE2, MYO2, CDC55, and NKD1 (DIP, 2002; <DIP Node: Calmodulin>.
The Database of Interacting Proteins is the creation of David Eisenberg at UCLA. This database presents protein interaction in a similar manner to Curagen's Pathcalling, but is not limited to S. cerevisiae. One of the interesting features of DIP is that it gives interaction information for a number of proteins in the form of a diagram showing nodes connected by lines. The interactions are putative, and the confidence in the interaction is indicated by the width of the bar connecting the nodes. When there are a large number of interactions to display, it can lead to some fairly indecipherable images, an example of which is calmodulin, as seen above. Thankfully, one is able to follow a link to a list of the direct interactions given for calmodulin. As expected for such a ubiquitous gene, the number of interactions listed amounts to 65. That list can be accessed by following this link: DIP protein = calmodulin. Some of the genes listed are CMK1, CMK2, and MYO2, all three of which are listed as known interactions by the Cyert paper cited in Assignment 2. In addition to these, there are a number of other myosin related proteins listed, as well as many other known and unknown genes.
MIPS:
The MIPS Comprehensive Genome Database provides a lot of information on S. cerevisiae, and has individual gene annotations which often complement information provided by SGD. In fact, I wish I had known about this while preparing my previous 2 websites. A search of MIPS gave a list of 12 protein interactions for calmodulin. They included the expected CMK1, CMK2, and MYO2. It also listed interactions between calmodulin and SLT2, MYO5, and NUF1, which were all noted by Curagen's Pathcalling. SLT2 is involved in signal transduction, MYO5 is involved in endocytosis and budding, and NUF1 is involved in connection of the spindle pole body to microtubules. All of these processes are known to include calmodulin as a consituent. MIPS also predicts the isolelectric point of calmodulin to be 4.01.
Y2H:
The Yeast Resource Center of University of Washington provides a comprehensive list of protein interactions for S. cerevisiae discovered using the yeast two-hybrid method of discerning protein interactions. Although CMD1 was not used in the first round of tests, the YRC's page of newer discoveries lists the following interactions:
|
Bait |
Prey |
CMD1 |
YML095C-A |
CMD1 |
YMR111C |
Both YML095C-A and YMR111C are unannotated ORFs.
Benno Schwikowski PDF file:
On the Genomics Place website, under chapter 6, there is a link to a PDF file which presents a figure from a paper that was a collaboration between Stan Fields, Peter Uetz, and Benno Schwikowski. The file is a diagram depicting 2,709 interactions among 2,039 protein. The figure shows protein names in color-coded boxes, connected by color-coded lines.
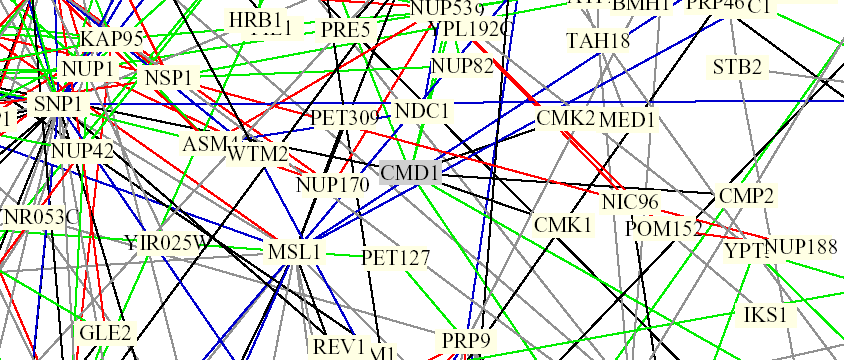
Figure 2. Screen capture from Benno Schwikowski PDF file depicting numerous protein-protein interactions. CMD1 can be seen in the middle, surrounded by a grey box which indicates the protein is involved in chromatin structure. It is connected by black lines to CMK1 and CMK2, as well as SNP1. Black lines are indicative of different cellular roles and localizations. The green lines projecting upwards from the grey box connect with MYO5, IQG1, and CNA1 (not visible here, left to right). Green lines indicate identical cellular roles but different localizations (The Genomics Place, 2002; <Benno Figure 1>).
According to SGD, IQG1 is involved in actin filament organization, SNP1 is involved in mRNA binding, and CNA1 encodes for a subunit of calcineurin, a protein which interacts very closely with calmodulin. It is involved in cell wall organization and response to mating signal.
PROWL:
The PROWL database had no useful hits for calmodulin. Strangely enough, one of the hits was a link to the NCBI entry for YBR108W, my unannotated ORF.
ExPASy:
The ExPASy database, which features information on protein analysis from 2D-PAGE, has no information on CMD1.
What is There?:
The What is There? site contains information on protein function and metabolic pathways. It contains no information on CMD1.
Experimental Design:
In addition to confirming a number of interactions revealed by pre-genomic and pre-proteomic methods, my exploration of calmodulin in yeast cells has shown a number of other previously unmentioned interactions or clusterings. Granted, these are putatitive, but it possible that calmodulin is involved in other pathways and processes previously unknown to us. This seems likely, considering that it is already known to be omnipresent throughout the cell. Therefore, I would like to carry out an experiment that might reveal some previously unknown interactions based on observations of the effects of a yeast mutant deficient for calmodulin's Ca2+ binding on other proteins. In assignment 2, I learned that calmodulin interacts very closely with 2 proteins: Cmk1p and Cmk2p, both of which are calmodulin-dependent protein kinases. According to the Cyert paper, mutants with CMD1 genes that are Ca2+ binding defective fail to active either of the protein kinases. Fortunately, this is conducive to study because, surprisingly, the phenotype is not lethal (2001). I would like to use a method developed by Brian Chait at Rockefeller University, in order to learn more about the chain of events brought on by calmodulin's activiation of calmodulin-dependent protein kinase 1 and 2. Two cell populations, one grown with regular copies of CMD1, and one defective for Ca2+ binding, would be grown in the presence of either 14N or 15N, both stable isotopes of nitrogen. These populations would then be combined, and the sample could be purified for a number of possible targets of the protein kinases. Next, analysis by mass spectrometry would reveal which proteins had been phosphorylated or not. The experiment would be repeated for cmk1p and cmk2p, individually. This method would possibly reveal known targets for these proteins as well as unknown targets, based on their relative levels of phosphorylation.
The Role of YBR108W in S. cerevisiae
As noted above, YBR108W is an unannotated ORF. It is located on the 2nd chromosome. Although little is known about this gene, my explorations have given me some information about its possible role.
DIP:
Figure 3. Screen capture of graph generated by DIP showing YBR108W node (red). This diagram is less complicated that that of calmodulin. One can see that DIP predicts 6 interactions for YBR108W (DIP, 2002; <DIP Node: YBR108W>). .
DIP predicts six direct interactions between YBR108W and other proteins, which I have summarized in table form below, using annotations from SGD:
Gene Encoding |
Function |
RVS161 |
cytoskeletal protein binding |
SEC27 |
ER to golgi protein trafficking |
LSM3 |
mRNA binding |
SEC21 |
ER to golgi protein trafficking |
COR1 |
ubiquinol-cytochrome c reductase |
LSB1 |
LAs17 Binding protein |
Another interesting thing that I found at DIP was that DIP lists YBR108W as a probable transcription factor.
MIPS:
MIPS lists YBR108W as similar to R. norvegicus atrophin-1 related protein. Although there are no specific entries at NCBI's Entrez, I did learn there that atrophin-1 is a gene related to dentatorubral-pallidoluysian atrophy, which is a neurodegenerative disorder. Beyond that, MIPS predicts the isoelectric point of YBR108W's protein to be 9.85. It lists one interaction with RVS161, one of the proteins observed in the DIP search.

Figure 4. Screen capture taken from MIPS page for YBR108W listing the interaction activity between YBR108W and RVS161/END6.
Y2H:
The Yeast Resource Center lists the following interaction, once again we see RVS161:
| Bait | Prey |
| RVS161 | RVS167 |
| YBR108W |
Benno Schwikowski PDF file:
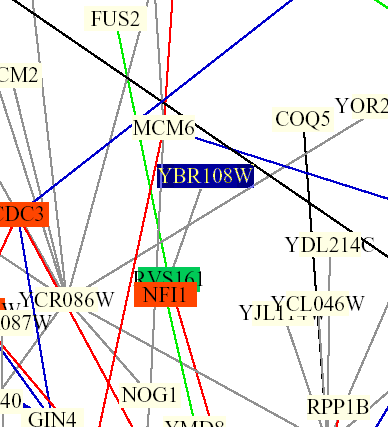
Figure 5: Screen capture from Benno Schwikowski PDF file depicting numerous protein-protein interactions (The Genomics Place, 2002; <Benno Figure 1>).
The only interaction of YBR108W depicted is with RVS161. The grey line indicates that localizations and cellular roles are unknown. The blue box indicates membrane fusion and the green box indicates cell structure.
PROWL:
A search of PROWL provided no information on YBR108W.
ExPASy:
A search of ExPASy provided no information on YBR108W.
What is There?:
A search of WIT provided no information on YBR108W.
Final Thoughts:
At the end of assignment three, the best guess I could make was that YBR108W might encode for a protein involved in organization of the cell wall. Now, the most recent exploration seems to point toward a relationship between YBR108W and RVS161 which encodes for a protein involved in cytoskeleton protein binding. However, this is confounded by 2 unqualified statements, that a) YBR108W encodes for a probable transcription factor or b) YBR108W encodes for a protein similar to atrophin-1 in R. norvegicus, made at DIP and MIPS, respectively. Neither DIP or MIPS provides any explanation for these listings, and I have not come across evidence for either in my past searches. I also went back to look through my webpage for assignment 3, to see if there was any listing of RVS161 amongst the microarray data. Alas, there was none. It would be interesting to see how the people who listed the interaction between YBR108W and RVS161 found it.
Experimental Design:
The fact that interactions have been observed and are documented in the databases above, I think, proves that YBR108W must indeed encode for a protein. According to the SGD entry, the protein that YBR108Wp is most often associated with, RVS161, is apparently required for survival after nitrogen, carbon, or sulfur starvation. The proteins encoded by RVS161 and several others form a complex that performs a number of regulatory activities following starvation or osmotic stress. SGD goes on to say that although RVS161 mutants are viable, mutation results in delocalization of actin cytoskeleton, high salt sensitivity, random budding pattern in diploid cells, defects in endocytosis, and reduced viability upon starvation. Based on the fact that DIP calls YBR108W a probable transcription factor, one might attempt to find evidence for this in the following way. If one would want to investigate if YBR108W encodes for a transcription factor involved in the regulation of RVS161 transcription, the first step would be to delete or mutate YBR108W (according to SGD, YBR108W mutants are viable). The next step would be to perform a northern blot, since the scope of this experiment is not expansive enough to require high-throughput. The northern blot would involve RNA taken from yeast cells with a deleted copy of YBR108W. Using a probe made up of the single-stranded DNA encoding for RVS161, you could measure the amount of RVS161 present in these cells. For the standard of comparison, one could perform the same northern blot with RNA from yeast cells with a working copy of YBR108W. For one final comparison, perhaps an extra copy of YBR108W could be inserted into the genome. If YBR108W does encode for a transcription factor, then one expect the northern blot to reveal less RVS161 in the YBR108W-lacking sample, and more RVS161 in the sample with two copies.
A simpler, more observation-based experiment would be to delete or mutate YBR108W in a number of samples of yeast cells. Then, these cells could be subjected to high salt or starvation. If YBR108W is involved in the transcription of RVS161, one would expect these cells to exhibit salt sensitivity and decreased viability following starvation. Two controls could be included with this experiment, one in which effect of high salt and starvation was observed in RVS161-deficient yeast cells, as well as high salt and starvation in wild type cells. In either this experiment or the one proposed above, if the final observations do not suggest a transcripion-based relationhip, then one must conclude that YBR108W must interact with RVS161 in some other way.