Databases Searched
Database of Interacting Proteins
Introduction to this Page
The purpose of this page is to use proteomic methods to determine what protein - protein interactions ADH1p is involved in. This is different than genomic methods that I used in the previous page " My Favorite Yeast Gene Expressions", where my conclusions were based all on microarray expression patterns of the yeast genome. For this page I am investigating the constantly fluctuating yeast proteome and determining the role(s) of ADH1p in yeast.
Database Search Results
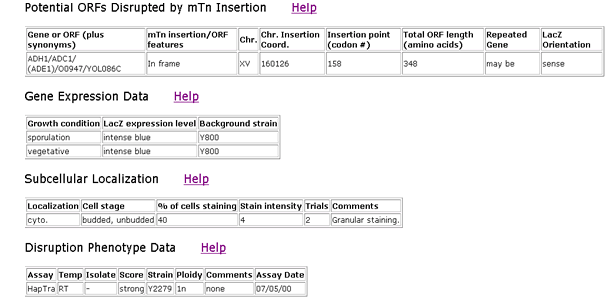
Triples Database

The Triples Database search showed that ADH1p is located in the cytoplasm. The LacZ gene was highly expressed when it was inserted into the ADH1 domain, which implies that ADH1p was being greatly expressed as well when the mTn was inserted into the ADH1 gene. This search also revealed that the HapTra phenotype was strongly exhibited when the mTn transposon was inserted. HapTra indicates that a cell is inviable in haploid transformants. Essentially the insertion of mTn into ADH1 results in an inviable cell. ADH1p is involved in the fermentation process and manufacturing of energy for the cell. It is reasonable that a disruption of the ADH1 gene would cause an inviable cell.
Database of Interacting Proteins
The nodes that the Database of Interacting Proteins ( DIP ) associated ADH1p ( red node in the middle ) with were not proteins that were not related to one another. The two closest known proteins to ADH1p were Leu2p and IMB4p that are proteins involved in leucine production and nuclear protein import respectively. There are no proteins in this diagram that are directly involved in fermentation or even energy production. This indicates that the DIP database does not have a very accurate model for protein - protein interactions. Although, I also noticed that there are no thick lines associating ADH1p with any of the other proteins, which means that the interactions shown on this diagram with ADH1 are weak and should not be taken with great confidence.
YRC Two-Hybrid Analysis
YRC Two-Hybrid Analysis yeilded no results for ADH1 protein interactions.
Munich Information Center for Protein Sequences
The MIPS database yielded one hit for a protein - protein interaction between ADH1p and another protein, which is TUB1p. TUB1p is a protein that is located in the cytoskeleton and is primarily involved in the mitotic cell cycle and cell cycle control. It is possible that ADH1p could interact with TUB1p since ADH1p is located in the cytoplasm. However, it is unclear why ADH1p and TUB1p would interact since ADH1p is involved in the fermentation process.
Scop Superfamily
A search of the Scop Superfamily amino acid sequence search revealed that ADH1p is part of the Alcohol Dehydrogenase superfamily. This finding is obvious because ADH1 is alcohol dehydrogenase. Though this search was not particularly useful, it provides reassurance that it works, which will be important for my search of my unknown yeast protein YOL085C.
Yale Gerstein Lab
Like the Scop Superfamily search, the results of the Yale Gerstein Lab were already well known. The results of this search can be shown HERE.
3Dpssm
The results of the amino acid sequence search for ADH1 were predictable. There were mutliple hits for alcohol dehydrogenase in humans. The protein that showed the best alignment with ADH1 was the human protein ADH2 which is found in human liver tissue.
What is There?
There were no results found for ADH1 in the WIT database.
Protein Interaction Files
This is the Membrane Protein Interactions file and it links ADH1 directly to TUB1, which is the same interaction that was found in the MIPS search. Likewise, the green color of ADH1 indicates that it is involved in cell cycle control. The two black boxes connected to TUB1 are BIM1 and ALF1, which are proteins involved in RNA turnover. The rest of the boxes have no other characterized interaction.
The other three protein interaction files had no charaterized interactions for ADH1.
Benno Figure1 § Aging § Degradation
Experimental Design
ADH1 is a well known gene and it's protein ADH1p is a common protein that has been studied extensively. ADH1p is an integral component of the fermentation process of yeast cells. In the MIPS search and the diagram shown in the Protein Interaction File, ADH1 is apparently a component of cell cycle control as well. To test whether an ADH1p/TUB1p interaction occurs two experiments could be done to confirm or disprove such an interaction. First, you could put a fluorescent tag on ADH1p and use microscopy to see if the ADH1 is found on the cytoskeleton of the cell. If the tagged ADH1 were found on the cytoskeleton, it would strongly indicate that there is a protein - protein interaction between ADH1p and TUB1p. Secondly, since the deletion of ADH1 results in a viable cell you could test to see a deletion of ADH1 would cause any abnormalities with cell cycle control or the mitotic cell cycle process. This test only proves that ADH1p has a significant role in cell cycle control or not. A deletion test would not confirm or disprove that there is an interaction between ADH1p and TUB1p.
Referecnces
Protein Interaction File. http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/seq/Benno/membrane.pdf Accessed 14 Nov 2002.
SGD. 2002. http://genome-www.stanford.edu/Saccharomyces. Accessed 15 Nov 2002.