*This webpage was created as an assignment for an undergraduate course at Davidson College*
Yeast Expression of the Non-Annotated LEE1: Using DNA microarray studies I investigated the possible function of LEE1. LEE1 is a non-annotated gene found in Saccharomyces cerevisiae. For more information on the possible functions of LEE1 please see my previous website.
Expression during sporulation: I first looked at the expression of LEE1 during sporulation. To see the abstract of the associated paper click here.
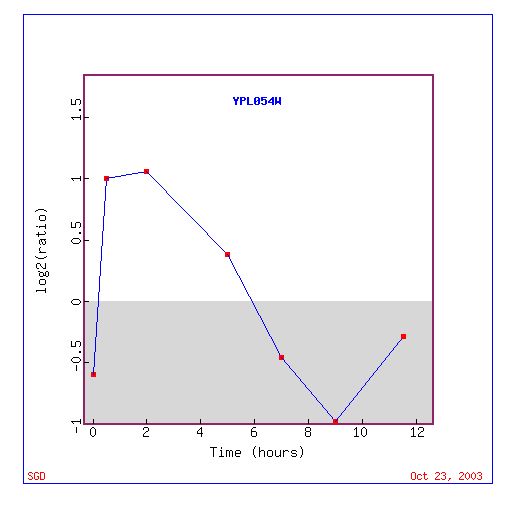
Figure 1: The expression levels of LEE1 over a period of time at a log base 2 ratio. (http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl?id=26598&dataset=sporulation&type=graph).
Figure one shows the levels of expression for LEE1 during sporulation at various time points. At the beginning of sporulation LEE1 is highly induced it then becomes repressed and then again begins to be induced toward normal levels.
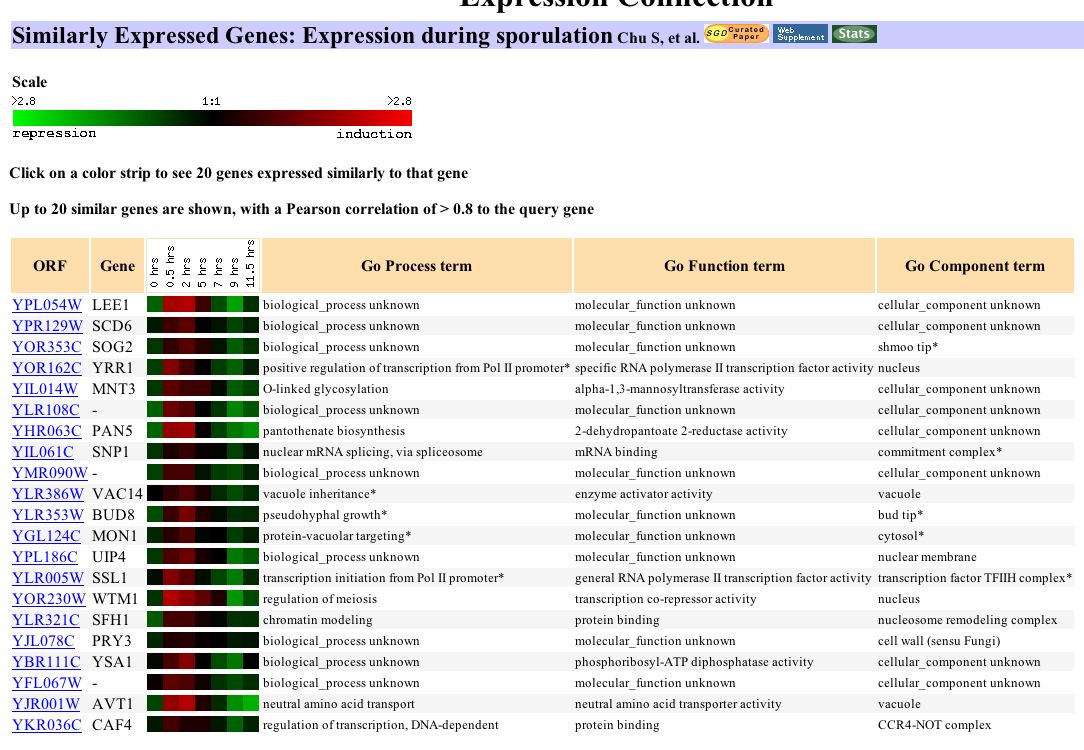
Figure 1: Association of LEE1 expression levels during sporulation to 23 genes with similar expression profiles. (http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl?orf=YPL054W&dataset=sporulation&type=similar).
This figure demonstrates that LEE1 associates with a variety of genes during sporulation. Many of these genes are unannotated and have no known biological function. CAF4 and YRR1 are invovled in regulation of transcription. SSL1 is a transcription initiator. WTM1 also has a similar expression profile and is involved in regulation of meiosis.
Diauxic Shift:
Next I examined the expression levels and associated genes of LEE1 during a diauxic shift. To read the abstract of the associated article please click here.
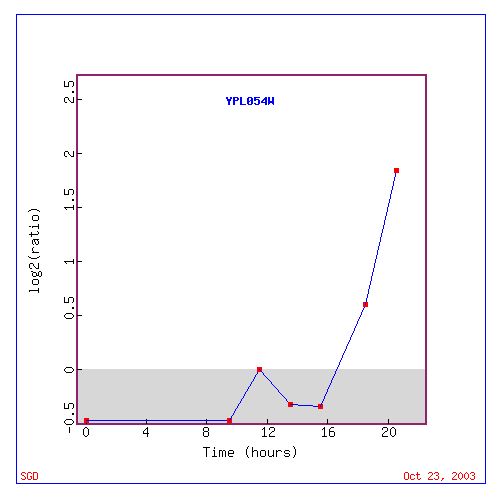
Figure 3: This figure shows the expression levels of LEE1 during the diauxic shift.(http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl?id=26598&dataset=diauxic&type=graph).
Based on figure three LEE1 appears to be repressed for the first 8 hours of the diauxic shift. It is then induced to 1:1 ratio and then repressed again. At 16 hours LEE1 is highly induced.
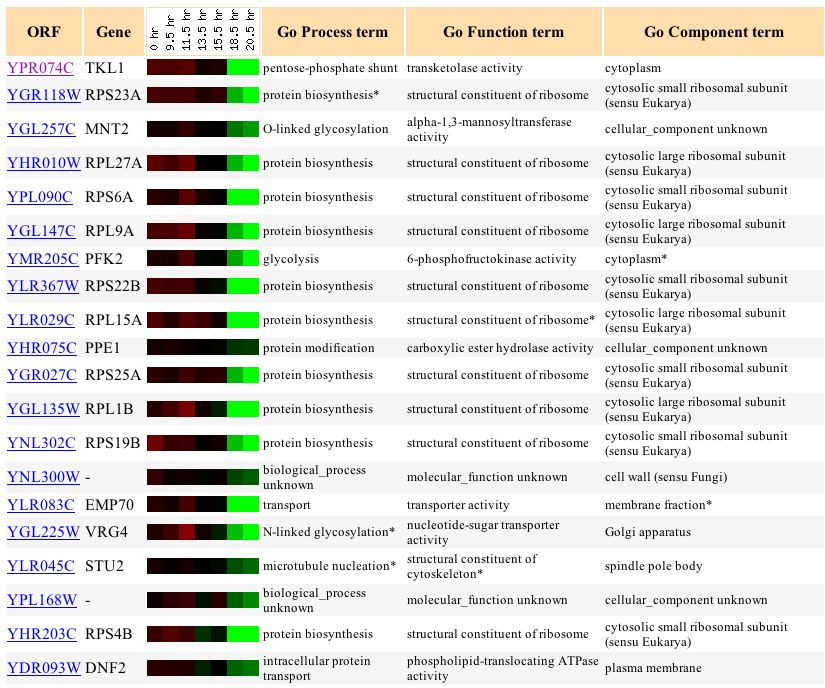
Figure 5: This image shows genes with similar expression levels to LEE1 during a diauxic shift. (http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl?orf=YPL054W&dataset=diauxic&type=similar).
According to figure 5 LEE1 has similar expression levels with some interesting genes. LEE1 is similar to GAL3 and SIP4 which are involved in regulation of transcription. There are also several similarities between LEE1 and genes for biosynthesis and metabolism. ACS1 is associated with acetyl -CoA biosynthesis has similar expression profiles as LEE1 along AGX1 which functions in the glyoxylate cylce and YAT2 which functions in alcohol metabolism.
Varying Zinc Levels: Based on my previous hypothesis that LEE1 was a zinc finger protein I examined the expression profile of LEE1 at varying zinc levels. To read the abstract of the associated paper please click here.
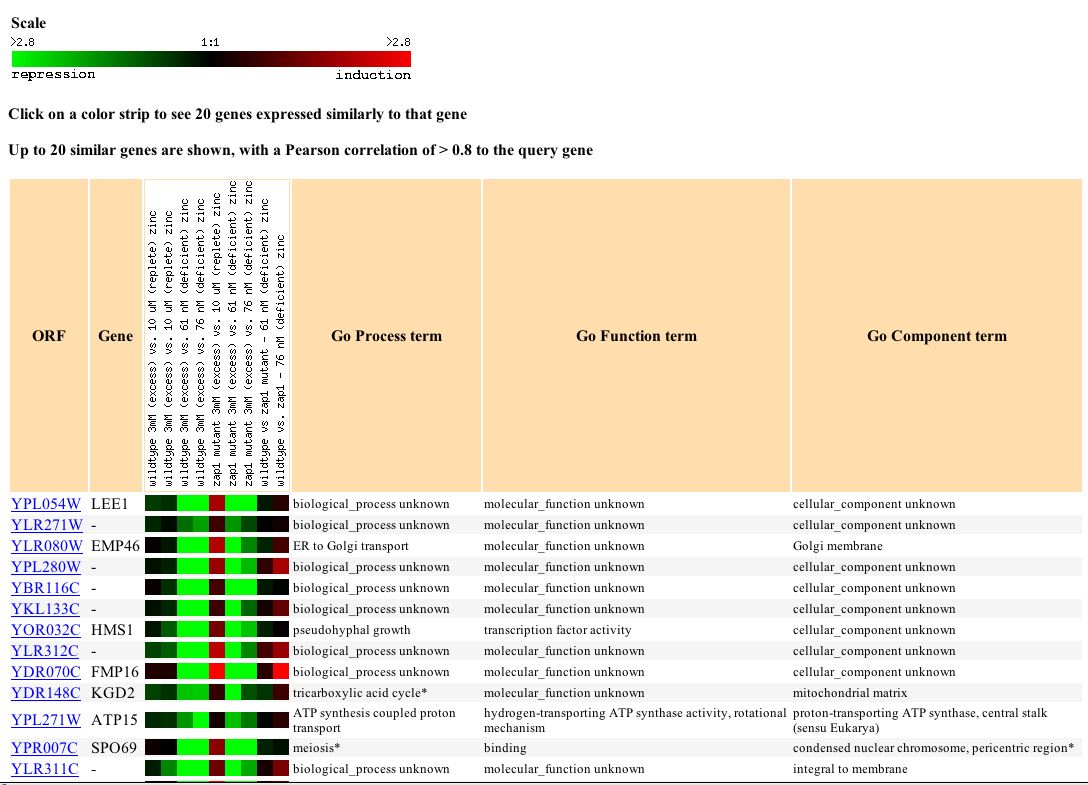
Figure 6: The expression profiles of genes with similar expression levels under varying zinc levels. (http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl?orf=YPL054W&dataset=zinc&type=similar).
Based on the information in figure 6 LEE1 associates with KGD2 which is involved in the tricarboxylic acid cycle and ATP15 which is involved in ATp synthesis proton transport. LEE1 expression profiles are also similar to EMP46 which is involved in ER to Golgi transport.
Conclusions:
My pervious investigation into the function of LEE1 lead me to hypothesize that LEE1 was a zinc finger protein. Zinc finger proteins have four amino acids that coordinate a zinc ion. These zinc finger motifs are often found in DNA or mRNA binding proteins. These proteins may help regulate transcription and translation. The transcription regulators CAF4 and YRR1 along with the transcription initiatior SSL1 have similar expression patterns to LEE1during sporulation. This supports the hypothesis that LEE1 may be a zinc finger protein which binds DNA and regulates transcription. This hypothesis is further supported by the association of the transcription regulators GAL3 and SIP4 with LEE1 duirng the diauxic shift. In contrast LEE1 expression patterns were not similar to any transcription regulators under varying zinc levels. In each of the three studies examined LEE1 had similar expression patterns to genes involved in biosyntheis and metabolism. These processes, however, do not support my previous hypothesis.
References:
Nelson, D. and M.M. Cox. 2003 Lehninger Principles of Biochemistry Thrid Edition. Worth Publishers.
Expression Connection 2003. http://genome-www4.Stanford.EDU/cgi-bin/SGD/expression/expressionConnection.pl.