this site is maintained by Nona Poulton under the supervision of Dr. A. Malcolm Campbell as part of the course requirements of Genomics at Davidson College, Davidson NC
Annotated vs Non-annotated Yeast Genes: RPB8 and YOR223W
Annotated Gene: RPB8
RPB8 encodes a protein that is a subunit for DNA-dependent RNA polymerase. RNA
polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside
triphosphates as substrates. Each class of RNA polymerase is assembled from
12 different polypeptides. The RNA polymerase II of the fission yeast Schizosaccharomyces
pombe consists of 12 Rpb subunits, of which four (Rpb1, Rpb2, Rpb3 and Rpb11)
form the assembly and catalytic core and five (Rpb5, Rpb6, Rpb8, Rpb10 and Rpb12)
are shared among RNA polymerases I, II and III. This subunit is shared by all
3 yeast RNA polymerases (called subunit H), making it very important to the
yeast genome and proteome (Sakurai et al, 2002).

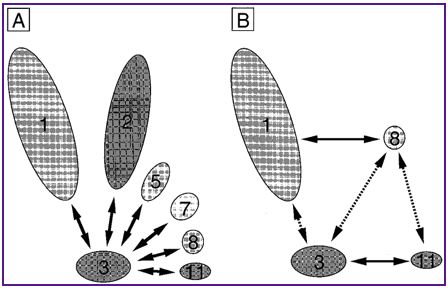
Figure 1: RPB8 is one of 12 subunits. The interactions of these
subunits have been worked out and can be seen above. (Permission pending, Kimura
et al, 2000, http://nar.oupjournals.org/content/vol28/issue4/images/large/gkd20406.jpeg).
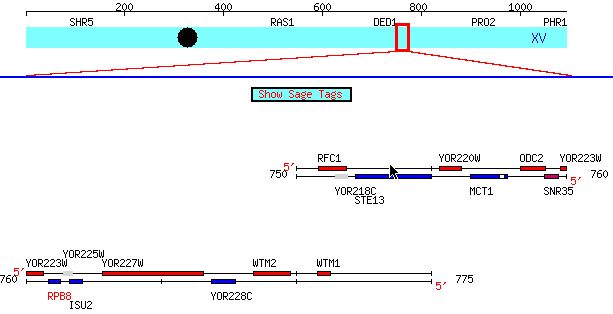
Figure 2: RPB8 is located on chromosome XV (it and its neighbors
are shown above). The image comes from SGD
acgtactccc aactgtggtc gcgctctcac cccttctgct gctctcgtgg ccccctcgcg
atggcgggca tcctgtttga ggatattttc gatgtgaagg atattgaccc ggagggcaag
aagtttgacc gagtgtctcg actgcattgt gagagtgaat ctttcaagat ggatctaatc
ttagatgtaa acattcaaat ttaccctgta gacttgggtg acaagtttcg gttggtcata
gctagtacct tgtatgaaga tggtaccctg gatgatggtg aatacaaccc cactgatgat
aggccttcca gggctgacca gtttgagtat gtaatgtatg gaaaagtgta caggattgag
ggagatgaaa cttctactga agcagcaaca cgcctctctg cgtacgtgtc ctatgggggc
ctgctcatga ggctgcaggg ggatgccaac aacctgcatg gattcgaggt ggactccaga
gtttatctcc tgatgaagaa gctagccttc tgaacctcgc ctgaagccag cctctctgcc
aagtcactca ggtcatgggc attgttcaag cctgagtggc agccgctctt gctcacctgt
tgaggaaggg ctggctcact gtccaccgtg gcggcatctt taactggcct ccactcaatg
ggaaactgac tcgcctgtga aagacacagt gggagagctg aaaatgaatc agaagcttta
tgtatatgat ttttaaatta aactttactt tttcagactg cccctaaaaa aaaaaaaaaaaaa
Figure 3: The nucleotide sequence of RPB8 (from NCBI's PubMed
search).
msntlfddif qvsevdpgry nkvcrieaas ttqdqckltl dinvelfpva aqdsltvtia
sslnledtpa ndssatrswr ppqagdrsla ddydyvmygt aykfeevskd liavyysfgg
llmrlegnyr nlnnlkqena yllirr
Figure 4: The amino acid sequence of RPB8 (from EntrezProtein).
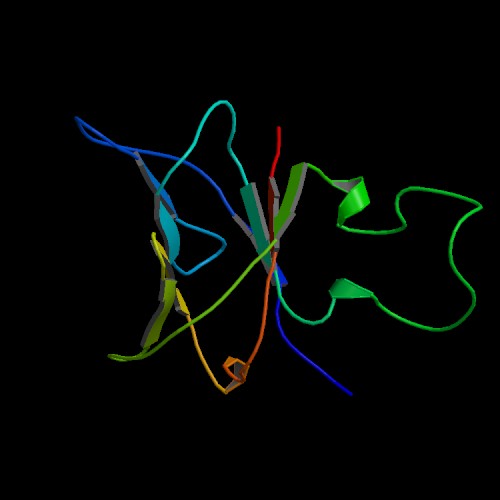
Figure 5: Crystal structure of RPB8 (from PDB),
showing RNA polymerase, subunit H.
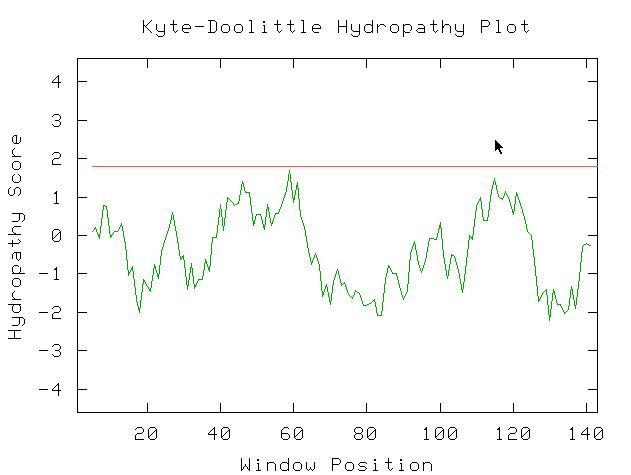
Figure 6: Kyte-Doolittle Hydropathy plot. The protein is not
a transmembrane protein, which would mean that there would have to be at least
one peak above 1.8 on the hydropathy score. It makes sense, however, when you
consider that its main function is RNA polymerase, which functions inside the
nucleus.

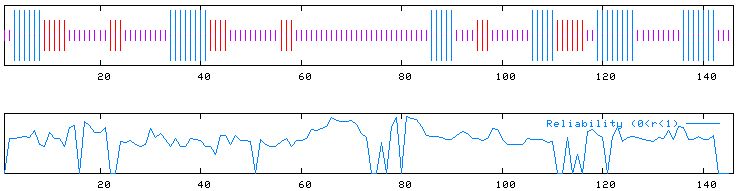
Figure 7: PREDATOR results. Predicts that there is a high frequency
of random coils in the protein product and upholds the data found in the Kyte-Doolittle
hydropathy plot for the proteins function (not an integral membrane protein).
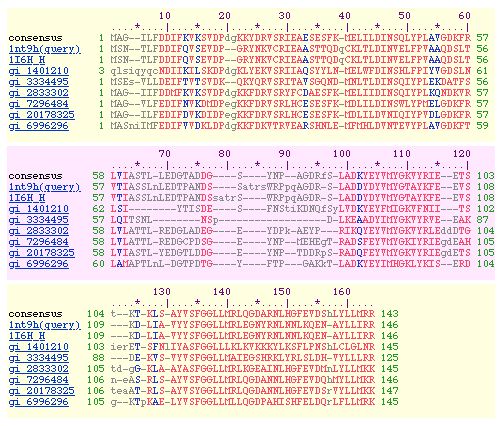
Figure 8: Homologues. The human homologue of RPB8 is POLR2H.
Other homologues can be seen above: gi_7296484 is the Drosophila homologue and
gi_6996296 is the Arabidopsis homologue. This gene is highly conserved between
species because it is crucial to protein production. This figure is courtesy
of the SGD website.
GO Information*:
Biological Process: Synthesis of RNA from DNA
template via RNA-polymerase (I, II, and III), of which RPB8 encodes a subunit.
Synthesis starts at the RNA-polymerase-specific promoter.
Molecular Function: As stated above, the biological function of this
protein is DNA-dependent RNA polymerase, where it functions as one subunit of
RNA polymerase I, II, and III. Here, it aids in the catalysis of DNA-template-directed
extension of the 3'- end of an RNA strand, one nucleotide at a time.
Cellular Component: RNA-polymerase initiates RNA synthesis from DNA
inside the nucleolus.
*GO information is from the SGD website for 'RPB8'
Nonannotated Gene: YOR223W
YOR223 is listed as an uncharacterized ORF on SGD. All GO information is listed
as unknown, and SGD describes the gene as a "hypothetical ORF'. A PubMed
search for the protein yields no results, and Entrez protein categorizes it
as an 'unknown protein'. Despite this lack of information, there is a lot we
can learn about the gene and its protein product. First I will consider its
location.
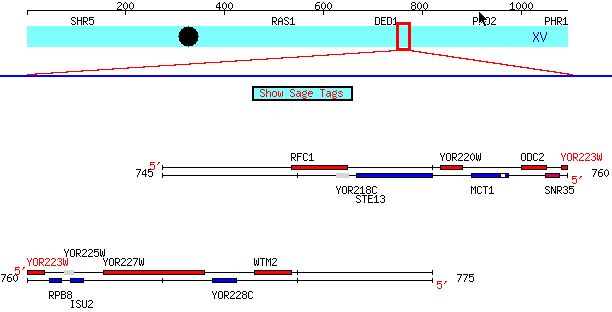
Figure 1: Location of YOR223W on Chromosome XV. It is located
on the Watson strand near RPB8 (above). The image comes from SGD.
msaepllpth ngsqggevrs pdqkfivirf sdvsvrdlql nisnvpfsni nthwlrrmcr
elrpqqtqkr rlkfirngsi lnthskiaee lthyfdtann snvatgtsva peqnnyyihc
iigteeltqa elanedlkdd atpsndsmtt qaigfdrlrs vgfteqeiel lrqqfratyg
dleeeeerla qngnrddegh dirqleeqwm esgsgtaqgn gagggnedrf nsvpianikh
nkdlllgicv gfffgvfgil lmkfdglfnr rqkmaifagv ivnvmfclvr gf
Figure 2: The amino acid sequence of YOR223W. This information
comes from EntrezProtein.
It is a larger protein than RPB8 (nearly 3 times its size), which is noteworthy
when considering its function (discussed later).
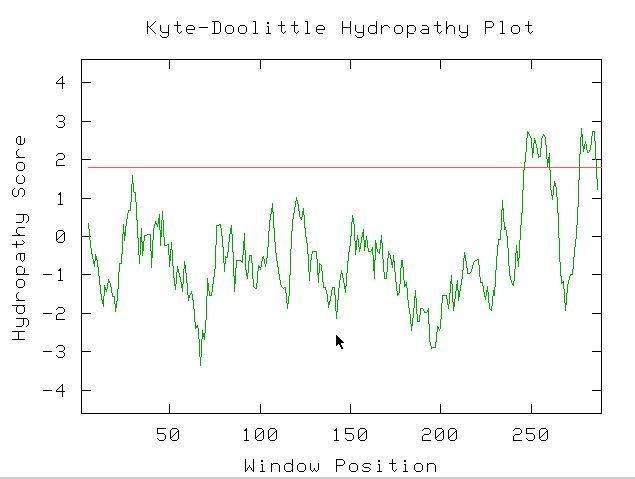
Figure 3: Kyte-Doolittle Hydropathy plot gives a good guess
at whether YOR223W is a transmembrane protein or not. The data suggest that
there is a transmembrane section because the hydropathy score is over 1.8. One
can infer from this data that the protein crosses the membrane twice.

Figure 4: PREDATOR results. The protein has many random coils and alpha helixes (53.8% and 34.9%, respectively).
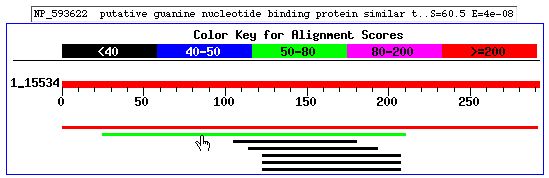
Figure 5: BLAST
results do not give proof of any conserved domains. But there is high sequence
simliarity to Schizosaccharomyces pombe guanine nucleotide binding protein,
seen in the figure above (the gloved hand is pointing to the gene of interest,
which comes from S. pombe). There are other sections that correspond to a variety
of other proteins (ranging from tRNA synthetase to protein phosphatase), which
makes it difficult to make any conclusions about YOR223W, however.
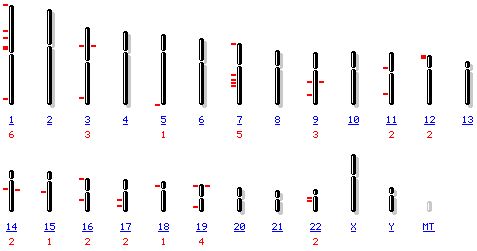
Figure 6: The above figure shows the results of a human map
viewer search for 'guanine nucleotide binding protein'. There are 32 different
guanine nucleotide binding proteins (that have been positively identified so
far) in the human genome, based on the data in this figure. None, interestingly
enough, show significant homology to YOR223W, the gene of interest (based on
BLAST2p searches).
GO Predictions:
Biological Process: Based on the information presented by Donzeauf
et al, YOR223W may be involved in the regulation of vegetative growth
and pseudohyphal development (the function of a 'G protein', or guanine nucleotide
binding protein).
Molecular Function: The function of the protein within the cell is
still unclear. It has been shown that the protein is not essential for cell
viability (from SGD
YOR223W page), however. It is most likely involved in growth and development,
however, if it is a putative guanine nucleotide binding protein.
Cellular Component: The protein is membrane-bound, as acheived from
the Kyte-Doolittle hydropathy plot results, thus it is probably in the membrane.
References:
Donzeauf, M and W Bandlow. (1999) The Yeast Trimeric Guanine Nucleotide-Binding
Protein α Subunit, Gpa2p, Controls the Meiosis-Specific Kinase
Ime2p Activity in Response to Nutrients. Mol Cell Biol.(9): 6110-19
Kimura, M and A Ishihama. (2000) Involvement of multiple subunit-subunit contacts in the assembly of RNA polymerase II. Nucleic Acids Res. 28(4):952-9.
Sakurai, H and A. Ishihama. (2002) Level of the RNA polymerase II in the fission yeast stays constant but phosphorylation of its carboxyl terminal domain varies depending on the phase and rate of cell growth. Genes Cells. (3):273-84.
Shpakovski, G. V.; Acker, J.; Wintzerith, M.; Lacroix, J. F.; Thuriaux, P.;
Vigneron, M. (1995)
Four subunits that are shared by the three classes of RNA polymerase are functionally
interchangeable between Homo sapiens and Saccharomyces cerevisiae. Molec.
Cell. Biol. 15: 4702-10.
Web References:
PDB
BLAST
PREDATOR
Kyte-Doolittle
Hydropathy Plot
Entrez and LocusLink
PubMed
SGD
© Copyright 2003 Department of Biology, Davidson College, Davidson
NC 28035
Send comments, questions, and suggestions to nopoulton@davidson.edu