This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Genes:
SPR3 and YGR058W
This web page follows two Saccharymyces cerevisiae genes, SPR3 and YGR058W. The two genes are located within close proximity to each other on chromosome 7. SPR3 is an annotated gene with a known gene product and function, so I will use information from web resources to elucidate the role of SPR3. YGR058W is a hypothetical open reading frame (ORF), or a non-annotated gene. Very little is known about YGR058W other than its sequence information, so I will use web tools in an attempt to characterize this unknown gene and its cellular role in yeast.
Annotated Yeast Gene: SPR3
The gene SPR3, also known as YGR059W, in Saccharomyces cervisiae encodes a septin protein. Septins are a group of proteins that are involved in cytokinesis, the organization of the cell surface during cell growth, and the localization of other proteins involved in cytokinesis.
In Gene Ontology terms, the molecular function of SPR3 is as a structural constituent of the cytoskeleton. The biological processes for SPR3 include "cell wall organization and biogenesis, cellular morphogenesis, cellular morphonesis during conjugation with cellular fusion, and assembly of the spore wall( sensu Fungi)". The cellular component for SPR3 is of the prospore membrane, the septin ring(sensu Saccharomyces), and the spore wall (sensu Fungi). (SGD, 2004; http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?sgdid=S000003291)
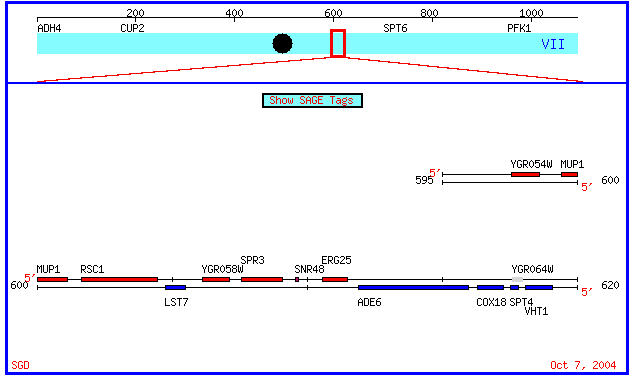
The gene SPR3 is located on chromosome VII from coordinates 607567 - 609105 in Saccharomyces cerevisiae (SGD, 2004; http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?sgdid=S000003291)
Though SPR3 plays a role in many important biological processes, yeast are viable without a functional copy of SPR3 and mitosis nor meiosis are affected by it's absence. However, there is a defect in sporulation if a functional copy of SPR3 is not present (H. Fares et al.,1996).

Figure 1. Section of S. cerevisiae chromosome 7 showing the location of both SPR3 and YGR058W.
SPR3 encodes a 512 amino acid that has a molecular weight of 59,845 Da (SGD, 2004; http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000003291).
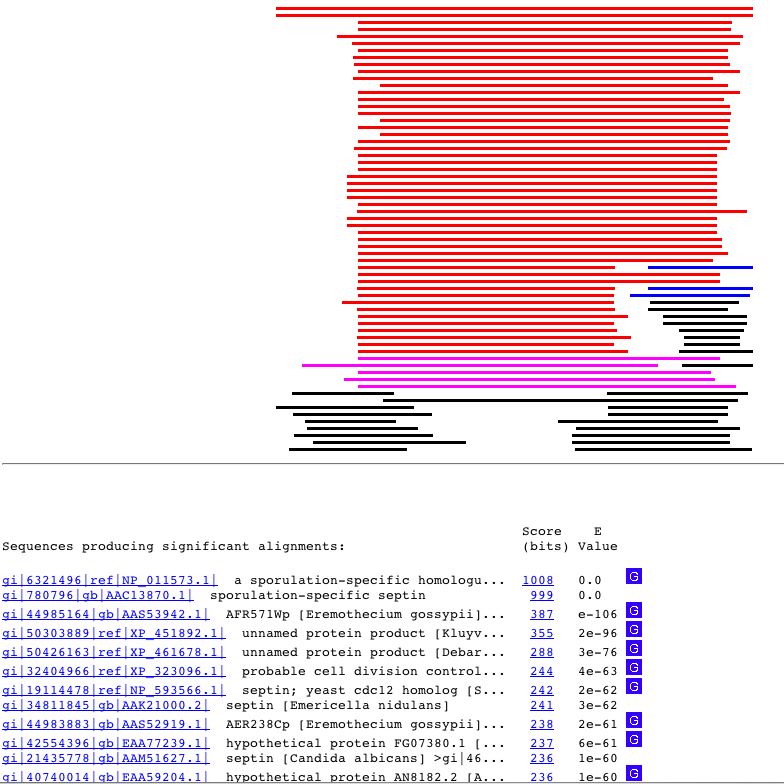
Figure 2. BLASTP results for S. cerevisiae SPR3 protein. Note that it does match up with the septin protein and is related to spororulation.To obtain results enter accession number NP_011573 into BLASTP to view.(NCBI, 2004)
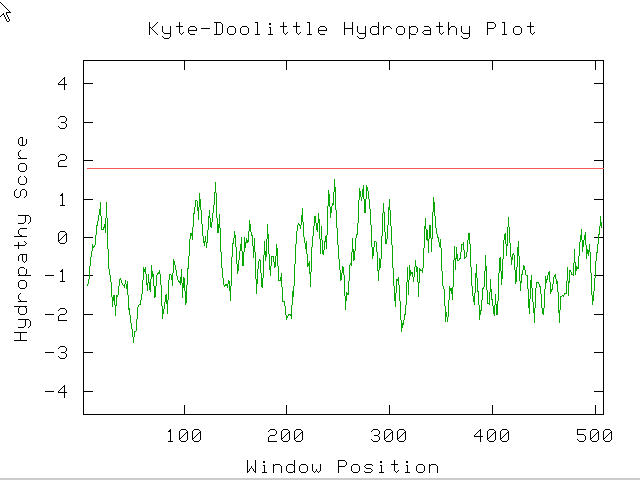
Figure 3. The protein product of SPR3 does not have a transmembrane region as determined by the Kyte-Doolittle Hydropathy Plot. If SPR3 did have an integral membrane domain than it would have a K-D hydropathy greater than 1.8, and notice above that it does not. (Kyte-Doolittle Hydropathy Plot; http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm)
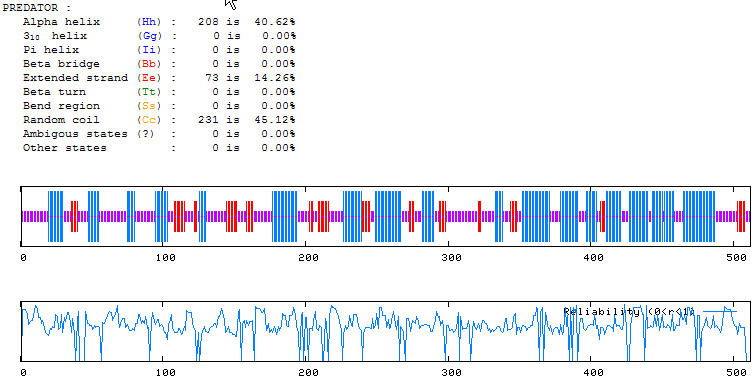
Figure 4. Predator results of SPR3 protein. These results indicate seemingly equal amounts of alpha helices and random coils within the secondary structure of the protein. (PREDATOR, 2004; http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html)
Homologues of SPR3 have also been found in Mus Musculus and Homo Sapiens. The human version is located at locus 1q21-q22 and is significantly shorter, 169 amino acids instead of yeast's 512. The human version's protein product has been found at elevated levels in tissues infected by human papillomavirus 11 and human papillomavirus 59 (Lehr et al, 2004).
Here are links to SPR3's protein sequence and nucleotide sequence.
Here's a link to the Entrez Gene entry for SPR3. This site contains links to PubMed articles, conserved domains, and many additional links associated with SPR3.
______________________________________________________________________________
Non-Annotated Yeast Gene: YGR058W
YGR058W is a hypothetical open reading frame(ORF) that is located in very close proximity to the SPR3 gene. It is also on the same strand as the SPR3 gene.The molecular function and biological process for this hypothetical gene's product are unknown, though the cellular component of this gene's product is in both the nucleus and cytoplasm. It is located on S. cerevisiae chromosome VII at coordinates 606140 - 607147(see Figure 1 above, YGRO58W is to the immediate left of SPR3) (SGD, 2004; http://db.yeastgenome.org/cgi- bin/locus.pl?locus=YGR058W). This gene's protein is predicted to contain 335 amino acids and have a molecular weight of 38,390 (SGD, 2004; http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000003290). Little is known about this ORF, so I will attempt to characterize it's molecular functions by using data from various online databases.
Genomic DNA Sequence of YGR058W: ATGTGTGCAAAGAAGCTCAAATACGCTGCTGGCGATGACTTTGTACGCTATGCTACACCT AAAGAGGCCATGGAGGAAACTAGACGTGAATTCGAGAAAGAGAAACAACGACAGCAACAA ATAAAGGTTACTCAAGCACAAACTCCCAACACTAGAGTCCACTCAGCTCCAATTCCCTTA CAAACTCAATATAACAAAAACAGAGCAGAAAACGGTCACCACTCCTATGGTTCTCCCCAA AGTTATTCTCCAAGACATACGAAAACACCTGTGGATCCTAGATATAATGTTATCGCACAG AAACCAGCAGGCAGGCCTATACCTCCAGCGCCAACCCATTATAACAACTTGAACACTTCC GCTCAACGGATAGCTTCCTCTCCTCCTCCCCTAATTCACAATCAAGCAGTGCCTGCACAA CTCTTGAAGAAAGTTGCACCTGCTTCGTTCGATAGCAGAGAAGATGTACGAGACATGCAA GTGGCCACACAGCTATTTCATAACCATGATGTAAAGGGCAAAAACCGACTGACAGCTGAG GAACTACAGAACTTACTACAAAACGACGACAACTCCCATTTTTGTATATCATCAGTAGAT GCGCTGATAAATTTATTTGGTGCTTCCAGGTTTGGCACTGTCAACCAGGCAGAATTCATC GCCCTATACAAAAGAGTGAAAAGTTGGAGAAAAGTTTATGTGGACAATGATATCAACGGA TCGCTCACCATTTCTGTAAGCGAATTTCATAACTCACTTCAAGAACTAGGATATCTAATA CCTTTTGAAGTTAGCGAGAAAACATTTGACCAATATGCTGAGTTTATAAACAGAAATGGA ACAGGAAAAGAACTAAAGTTTGATAAATTCGTTGAGGCGTTAGTTTGGCTAATGAGATTA ACAAAATTATTCAGGAAATTCGATACTAATCAAGAAGGCATTGCAACCATACAGTACAAA GATTTTATCTATGCTACATTATATTTAGGTCGTTTCCTACCTCATTGA
(SGD, 2004; http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YGR058W)
Amino Acid Sequence of YGR058W: mcakklkyaagddfvryatpkeameetrrefekekqrqqqikvtqaqtpntrvhsapiplqtqynknrae nghhsygspqsysprhtktpvdprynviaq kpagrpippa pthynnlntsaqriassppplihnqavpaq llkkvapasf dsredvrdmqvatqlfhnhdvkgknrltae elqnllqndd nshfcissvdalinlfgasrfgtvnqaefi alykrvkswrkvyvdndingsltisvsefhnslqelgylipfevsektfdqyaefinrngtgkelkfdkf vealvwlmrltklfrkfdtnqegiatiqyk dfiyatlylg rflph (NCBI, 2004; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?dopt=GenPept&cmd=Retrieve& db=protein&list_uids=6321495).
A BLASTP search(using accession number NP_011572) for this ORF reveals a large amount of similarity with other proteins (notice the hits with other low E-Values). Two of the proteins (the bottom two on Figure 1), have low E-Values of 2E-12, and are proteins whose entire functions are not known, but are known to be found in the cytosol and are involved in apoptosis in humans and mice respectively.

Figure 5. BLASTP search for YGR058W. Notice the large amounts of similar proteins, however also notice many of they are predicted and hypothetical proteins. (NCBI, 2004; http://www.ncbi.nlm.nih.gov/blast)
I then accessed several databases which allowed me to search for homology with other genes and to determine the degree of similarity between homologous genes.
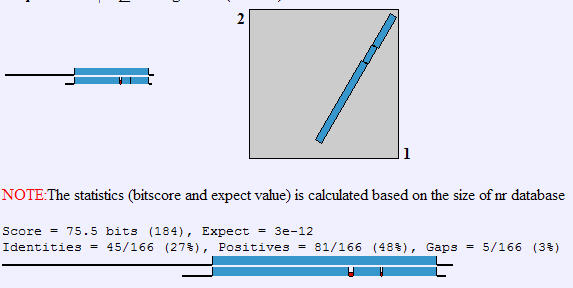
Figure 6. BLAST2 search between YGR058W and NP_035181(accession number for programmed cell death 6 protein in Mus Musculus). Important values to notice are the Postives and the Expect Value. The positives value of 48% indicates that percent of the amino acids are similar in quality(polar,non-polar,etc.) if not identical, and the low E-value of 3e-12 indicates that one would not expect these proteins to be so similar without being homologous.(NCBI, 2004; http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html)

Figure 7. HomoloGene results for YGRO58W. Note that only one gene was found to be homologous in this database. Further research from NCBI indicates that the E. gossypii gene is hypothetical as well. (NCBI,2004; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene&cmd=search&term=YGR058W)
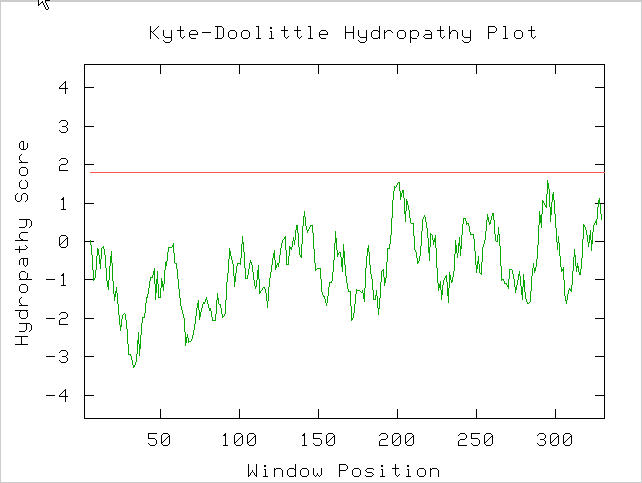
Figure 8. Kyte-Doolittle Plot of YGR058W. Note as was the case for SPR3, no peak goes above the 1.8 threshold, therefore there is no integral membrane region in the hyptothetical YGR058W protein.(Kyte-Doolittle Hydropathy Plot; http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm)
The Kyte-Doolittle hydropathy results correspond with this gene's cellular component of being in the nucleus and cytoplasm. That is, there is not integral membrane protein, so it cannot function as a extracellular transport or receptor protein.
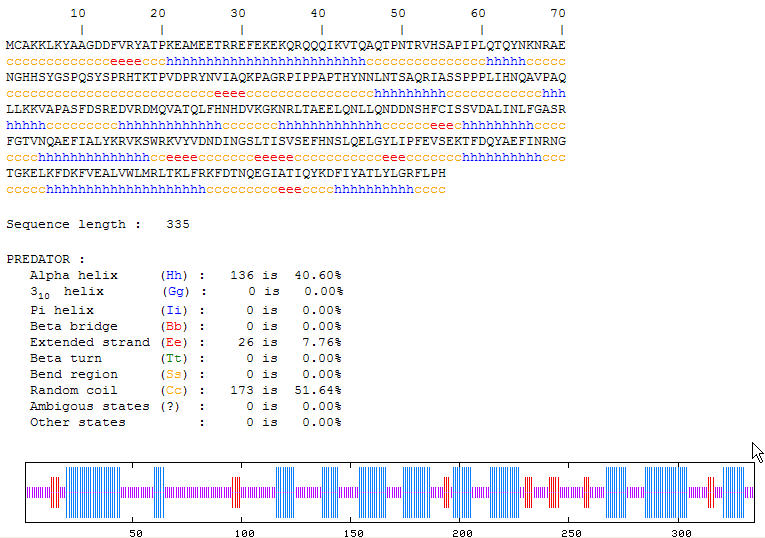
Figure 9. Predator results for YGR058W. These results indicate that the secondary structure consists mainly of alpha helices and random coiling. (PREDATOR, http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html)
Additionally, I found that it had a conserved domain that had a calcium binding motif(see Figure 10 below). Interestingly, the ability to bind to calcium ion may change the conformation of the YGR058W protein, which would seemingly allow it to carry out various other functions.

Figure 10. Conserved Domain results of YGR058W. The description says that this gene's product has an interaction with calcium.(NCBI, 2004; http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=14789)
Conclusion
SPR3 is a gene in Saccharomyces cerevisiae that produces a septin protein, and its molecular function, biological process, and cellular component are already documented as shown above. YGRO58W, however is a hypothetical ORF in the yeast genome. Its cellular component had already been localized to both the nucleus and the cytoplasm, but neither the molecular function or biological process are known. With the aid of many databases, I was able to determine some further information about the gene's functions. I further confirmed it's location within the cell itself with the Kyte-Doolittle results, and through BLASTP found that it's closest homologue(with a known function) is the cell programmed death 6 protein found in both mice and humans. From this information the best prediction I can make about this gene's functions are that it has a direct role in binding to calcium ions, which is associated with a conformation change in it's protein product that might allow it to carry out another function possibly related to the process of apoptosis. Though I can't be certain about this potential gene's functions, it is pretty amazing that I could hypothesize so much about what interactions this gene might be involved in simply by the use of these genomic databases.
Works Cited
Fares H, Goetsch L, Pringle JR (1996) Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol 132(3):399-411
Kyte-Doolittle Hydropathy Plot. 2004. <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm>. Accessed 8 Oct, 2004.
NCBI (National Center for Biotechnology Information). 2004. <http://www.ncbi.nih.gov> Accessed Oct. 8, 2004.
PREDATOR. 2004. <http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html> Accessed Oct.8, 2004.
SGD (Saccharomyces Genome Database). 2004. <http://www.yeastgenome.org/>. Accessed 8 Oct, 2004.
______________________________________________________________________________
Questions, Comments? E-mail John