*This page was created as an undergraduate assignment for a Genomics course at Davidson College*
YNK1 and YKL069W
My Favorite Yeast Gene Expressions
This webpage will be dedicated to displaying and discussing some of the expression patterns for YNK1 and YKL069W under different conditions and life cycles. YNK1 is a gene that encodes Nucleoside diphosphate kinase, a protein envolved in DNA biosynthesis (Jong and Ma, 1991). YKL069W is a non-annotated gene, with no known molecular function, or cellular component, or related biological processes. Based on sequence analysis only I have predicted that YKL069W is an enzyme found in the cytoplasm, and may be a part of a signalling pathway, or it could be a phosphodiesterase, based on its GAF domain. For more information on these two genes click here to view my previous webpage.
Expression Analysis of YNK1
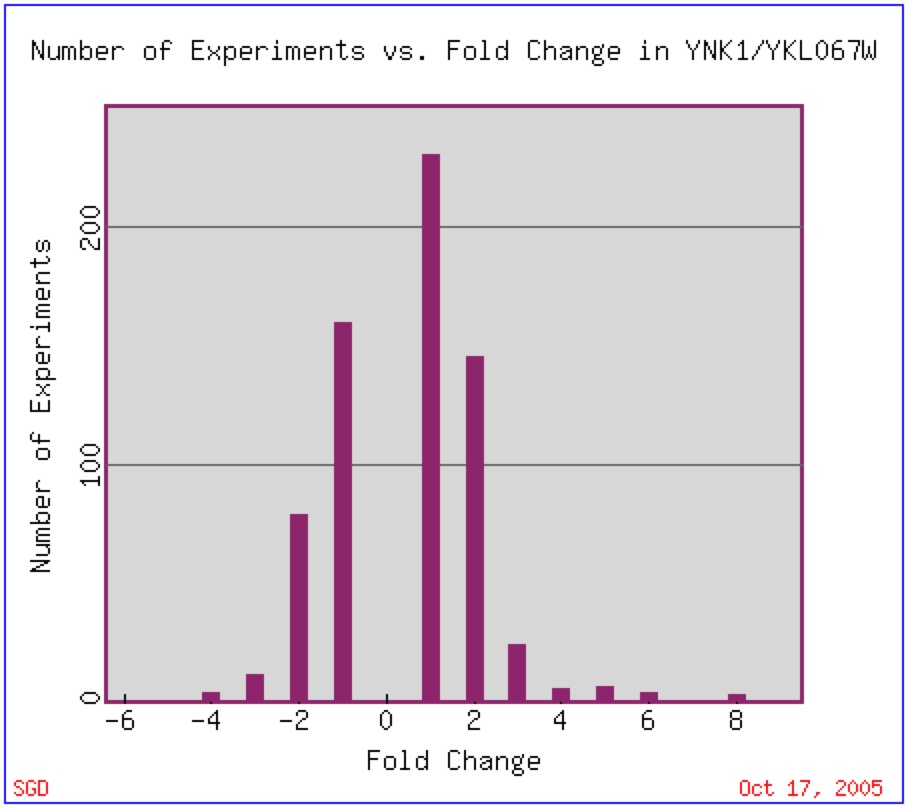
YNK1 expression has been found to vary little as the majority of experiments done have shown that a two-fold change in expression (repression or induction) is found most often (figure 1).

Figure 1: Expression Histogram for YNK1. This figure shows the number of experiments that have been done in which YNK1 mRNA changed by a number of fold changes (2, 4, 6, etc.) The positive number represent an induction while the negative number represent a repression. Image found at Expression Connection. Permission Pending.
YNK1 is found to have three fold or higher induction or repression in five different experimental conditions. During the time in which DNA-damaging agents are present there is a 3.3 fold induction in YNK1. This is not suprising as YNK1 encodes a protein involved in the biosynthesis of DNA (Figure 2).
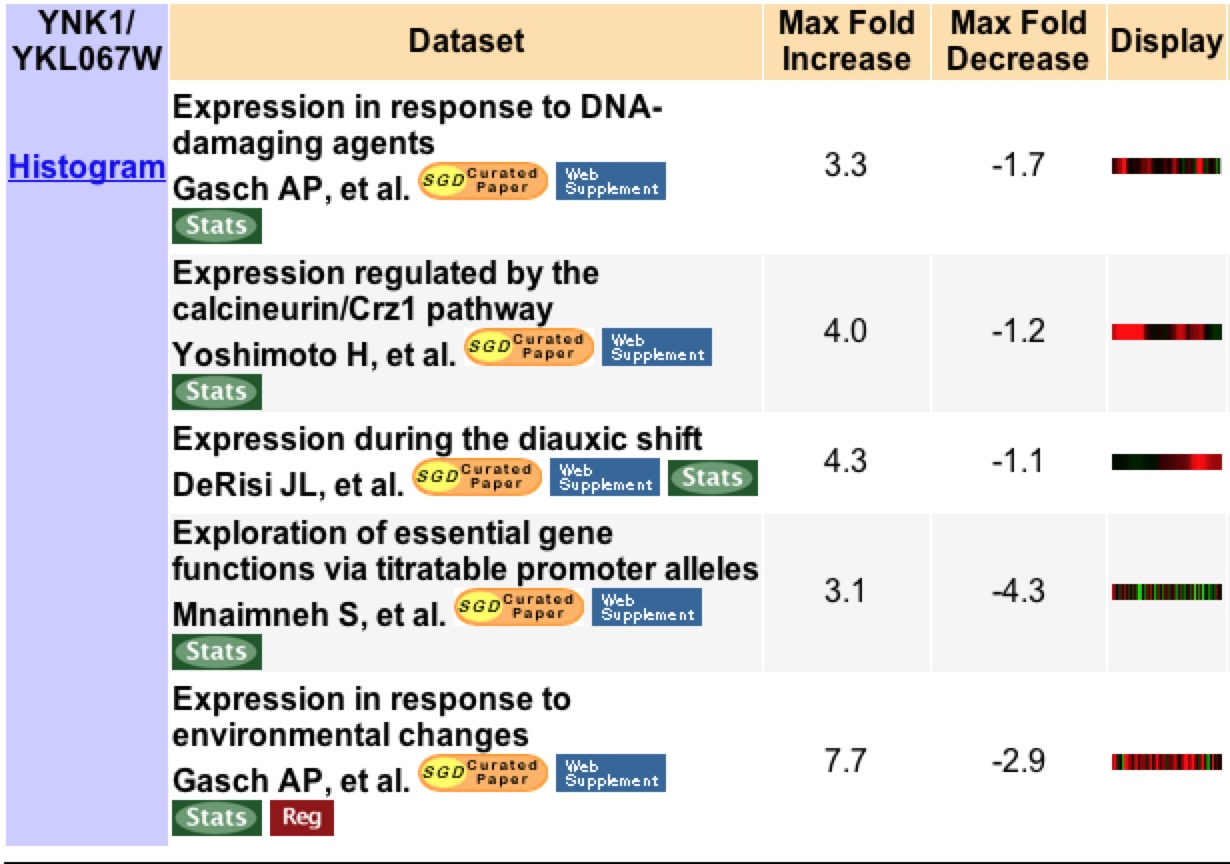
Figure 2: This image shows a list of all experiments that resulted in a 3 fold or higher expression change for YNK1 under various experimental conditions. Image from Expression Connection. Permission pending.
A four-fold induction of YNK1 also occurs during Ca induction of calcineurin, a transcription factor that is known to control a wide range of genes involved in "signaling pathways, ion/small molecule transport, cell wall maintenance, and vesicular transport" (Yoshimoto et. al 2002) (figure 3). Therefore one expects YNK1 to be involved in one of these areas.
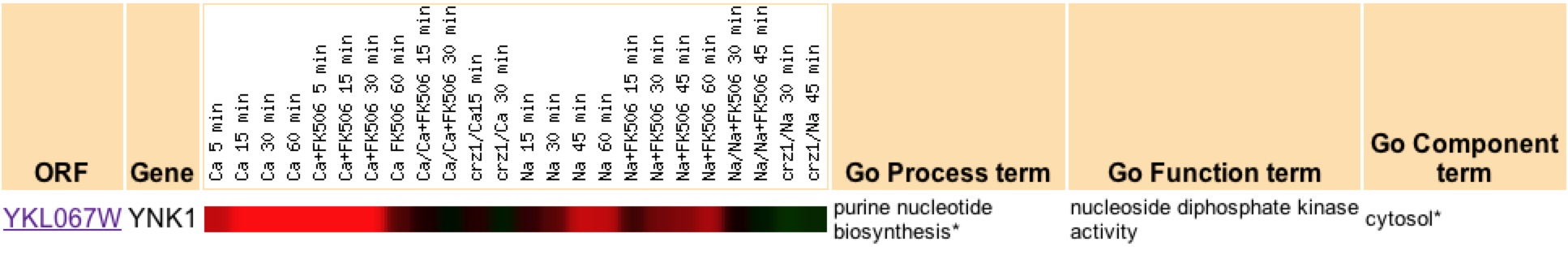
Figure 3: Expression of YNK1 during the calcineurin/Crz1 pathway. Induction of YNK1 occurs under the induction of calcineurin, a transcription factor. Image from Yoshimoto et. al 2002. Permission pending.
During diauxic shift YNK1 is induced four-fold suggesting that it is involved in energy metabolism, as genes that as induced during a diauxic are often related to energy metabolism (DeRisi et. al 1997) While the major function of YNK1's protein is in purine nucleotide biosynthesis, it is also believed that it may also phosphorylate other proteins, which could be directly involved in energy metabolism (Amutha and Pain, 2003).
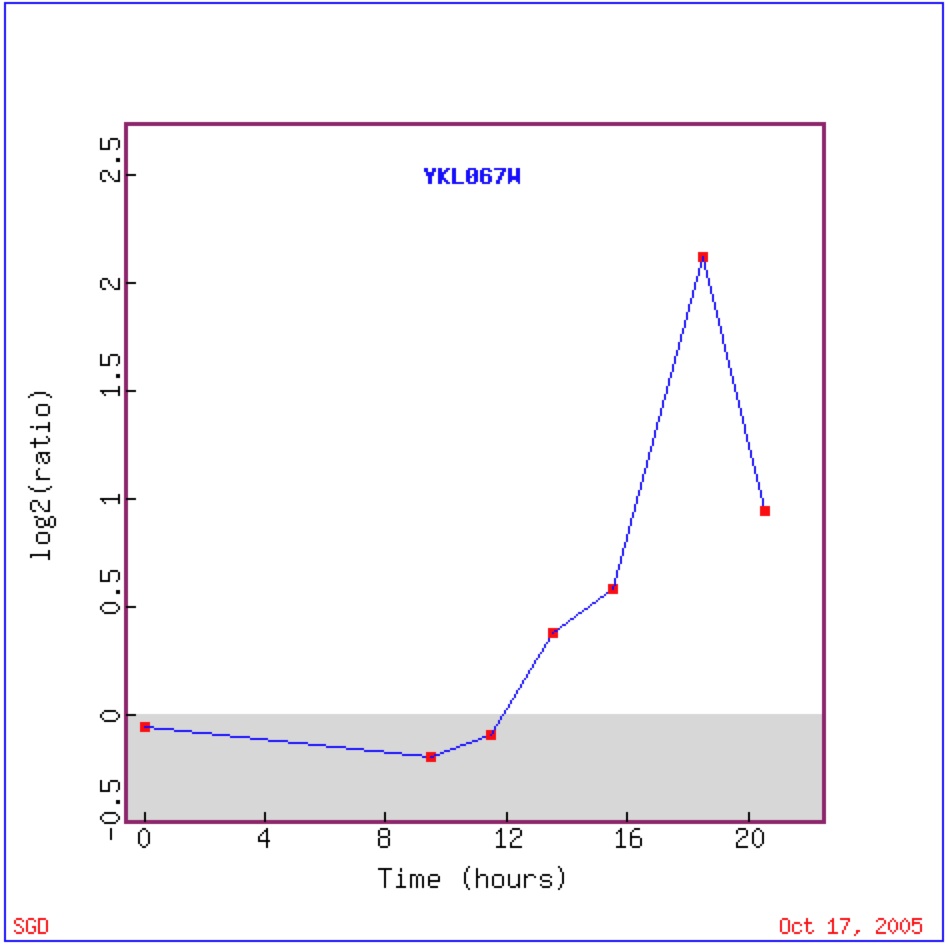
Figure 4: This image shows the expression of YNK1 over time during diauzic shift. For the first 20 hours YNK1 is repressed, during hour 12 YNK1 is induced and continues to be induced until time 18.5 hours, from here YNK1 begins to be repressed. Most likely returing to its original repressed state. Image from DeRisi 1997. Permission pending.
During the stationary phase YNK1 experiences up to a 7.7 fold induction (figure 5). The stationary phase in yeast occurs when there is no net change in the number of cells in a culture. This indicates that while replication is not taking place YNK1 is still being induced suggesting that the conversion of nucleosides diphosphates to nucloside triphosphates is needed to maintain cells during the stationary phase.

Figure 5: During the Stationary Phase YNK1 is induced up to 7.7 fold. After 2-4 hours YNK1 is induced, then expression levels return to normal after 5-7 days. Gasch et. al 2000. Permission pending.
Expression Anaysis of YKL069W
YKL069W expression has been found to vary greatly, although the majority of experiments that have been done have shown that a two-fold change in expression (repression or induction) is found most often (figure 6). One experiment that has been conducted shows a very high respression of YKL069W.
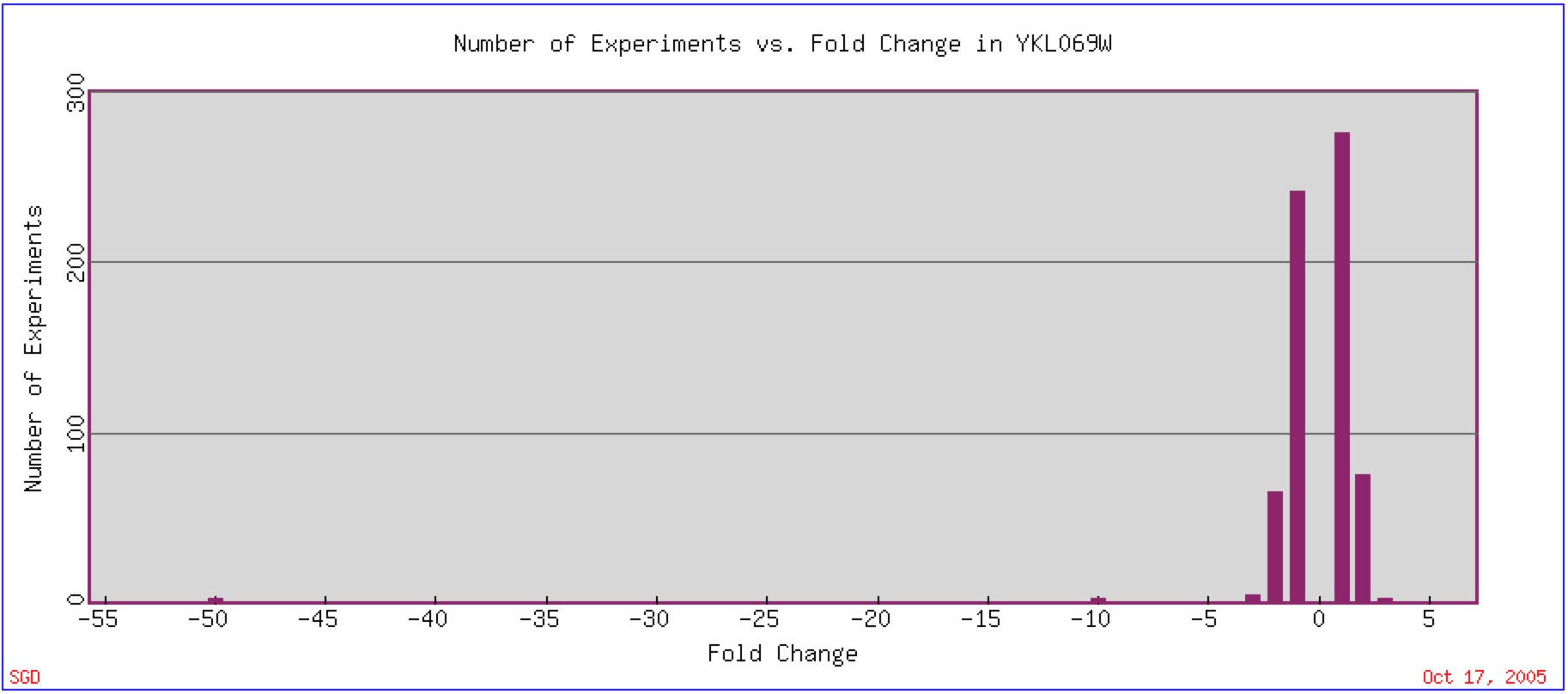
Figure 6: Expression Histogram for YKL069W. This figure shows the number of experiments that have been done in which YKL069W mRNA changed by a number of fold changes (2, 4, 6, etc.) The positive number represent an induction while the negative number represent a repression. Image found at Expression Connection. Permission Pending.
Of the experiments done to monitor YKL069W expression only two have produced more than a three fold change in induction or repression (Figure 7). I will focus on these experiments to make a better hypothesis on the function of this unknown gene.
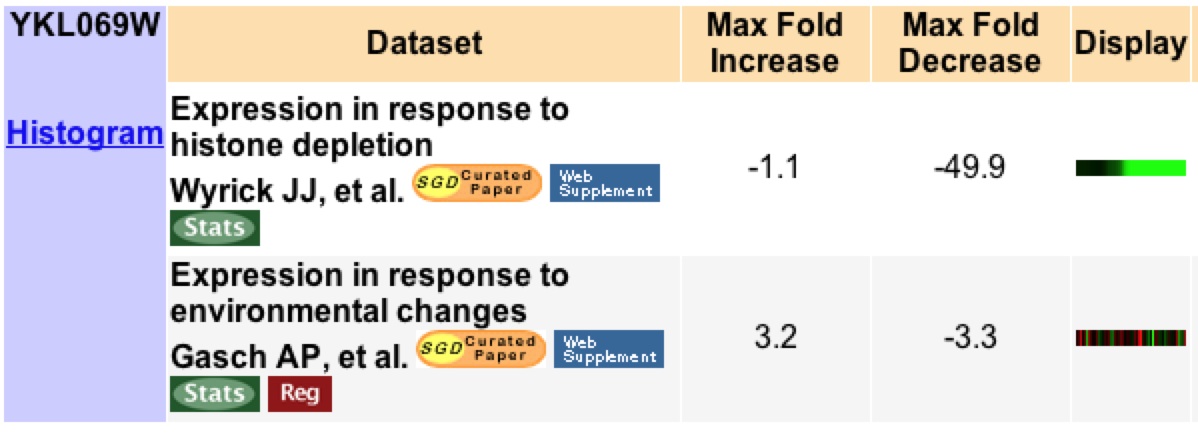
Figure 7: This image shows a list of all experiments that resulted in a 3 fold or higher expression change for YKL069W under various experimental conditions. Image from Expression Connection. Permission pending.
As nitrogen is depleted YKL069W becomes induced, suggesting that in the absence of nitrogen YKL069W is asked to function at a higher level (figure 8). Suggesting that its role possibly has something to do with nitrogen fixation or it is gene expressed in order to free nitrogen from its fixed sources in the cell for necessary cellular functions.

Figure 8: Expression profile for YLK069W under depleting nitrogen conditions. The longer nitrogen is withheld from the yeast the higher the induction of YKL069. Gasch et. al 2000. Permission pending.
YKL069W become repressed during the stationary phase after 2 days (figure 9). This suggests that because the cells are not dividing YKL069W's protein product is not needed while the cell is not reproducing or for cell maintenance.

Figure 9 Expression profile of YKL069W during the stationary phase. The longer the stationary phase the more repressed YKL069W becomes. Gasch et. al 2000. Permission pending.
Figure 10 displays the expression profiles of YKL069W under heat and cold shocks. It shows that during cold shock YKL069W is being repressed while during heat shock the opposite is true as YKL069W is being induced (figure 10).
Figure 10: Expression of YKL069W during heat/cold shock. During a cold shock YKL069W in repressed, but during a heat shock it is induced. Gasch et. al 2000. Permission pending.
The expression profile of YKL069W under environmental stresses was compared to all other expression files and no other genes were found to have similar expression profiles for the battery of conditions in these experiments (figure 11).

Figure 11: Expression profile of YKL069W compared to other gene expression profiles under similar conditions. There are no genes that have matching profiles. Gasch et. al 2000. Permission pending.
Under histone depletion YKL069 is highly repressed, a 49.9 fold decrease in production. Histones are proteins which DNA is coiled around to produce nucleosomes. During histone depletion most genes are induced due to more availability for transcription and translation of the genes when they are not bundle in nucleosomes (Wyrick et. al 1999) When histones become depleted the genes involved in protein biosynthesis, specifically in the ribosome, become highly repressed (figure 12). Based on the fact that 10 of the closest 20 expression patterns are for genes related to protein biosynthesis, I would say through Guilt-by-association that YKL069W is involved in protein biosynthesis and is likely related to a ribosomal subunit.
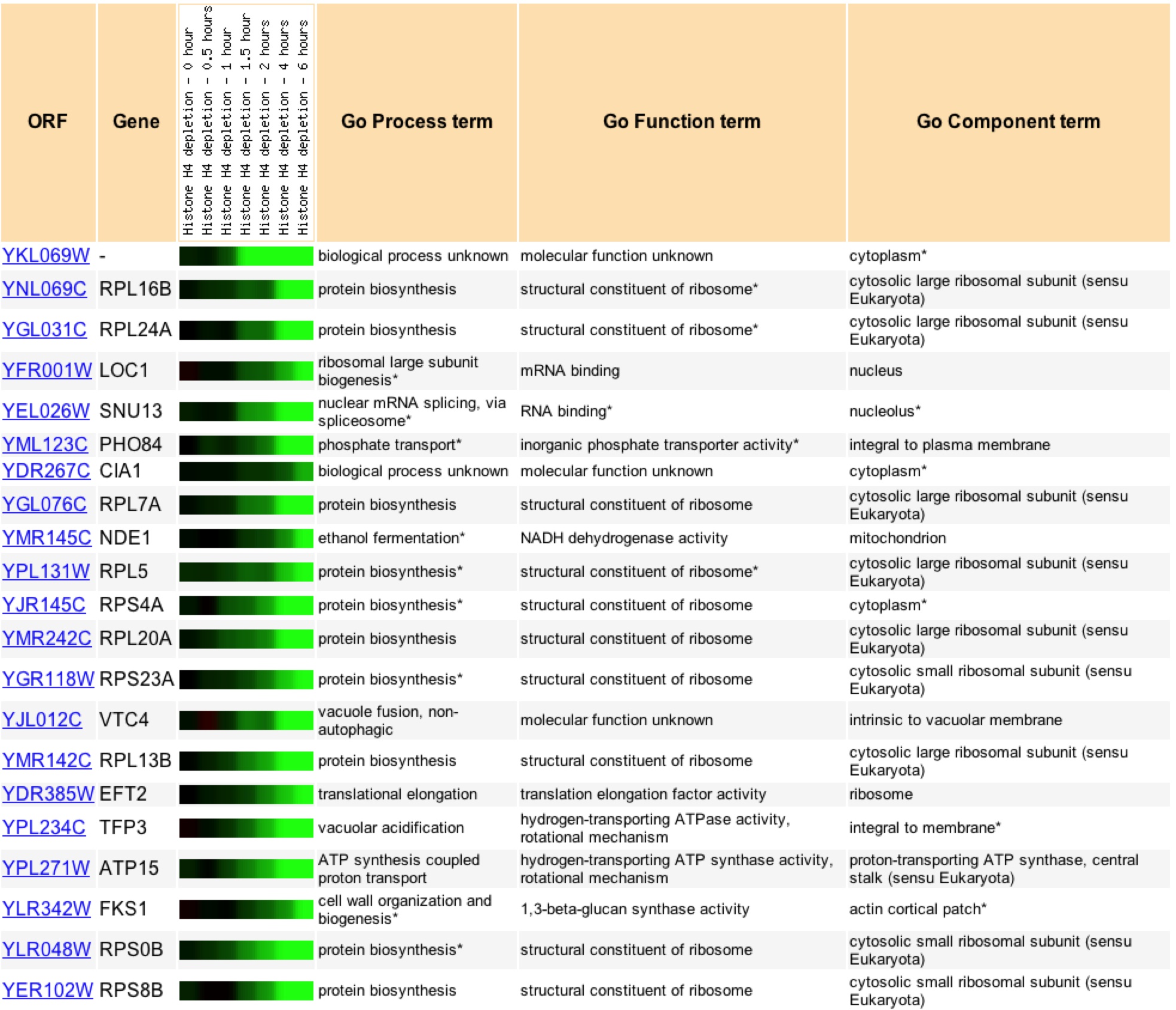
Figure 11: Expression profiles for YKL069W and its 20 closest profile matches for histone depletion, pearson correlation value of >.8 to YKL069W. Image found at Expression Connection. Permission Pending.
A search of Gene Ontology Term finder during the depletion of histones returned these results (figure 12). It is interesting to note that the highest clusterng frequencies occur during cellular physiology and physiological process, as well high frequecies of clustering with biosynthesis genes. Upon further searching one can find that the majority of the genes that have similar expression patters are found to be ribosomal proteins suggesting that through Guilt-by-association that YKL069W is a ribosomal protein.
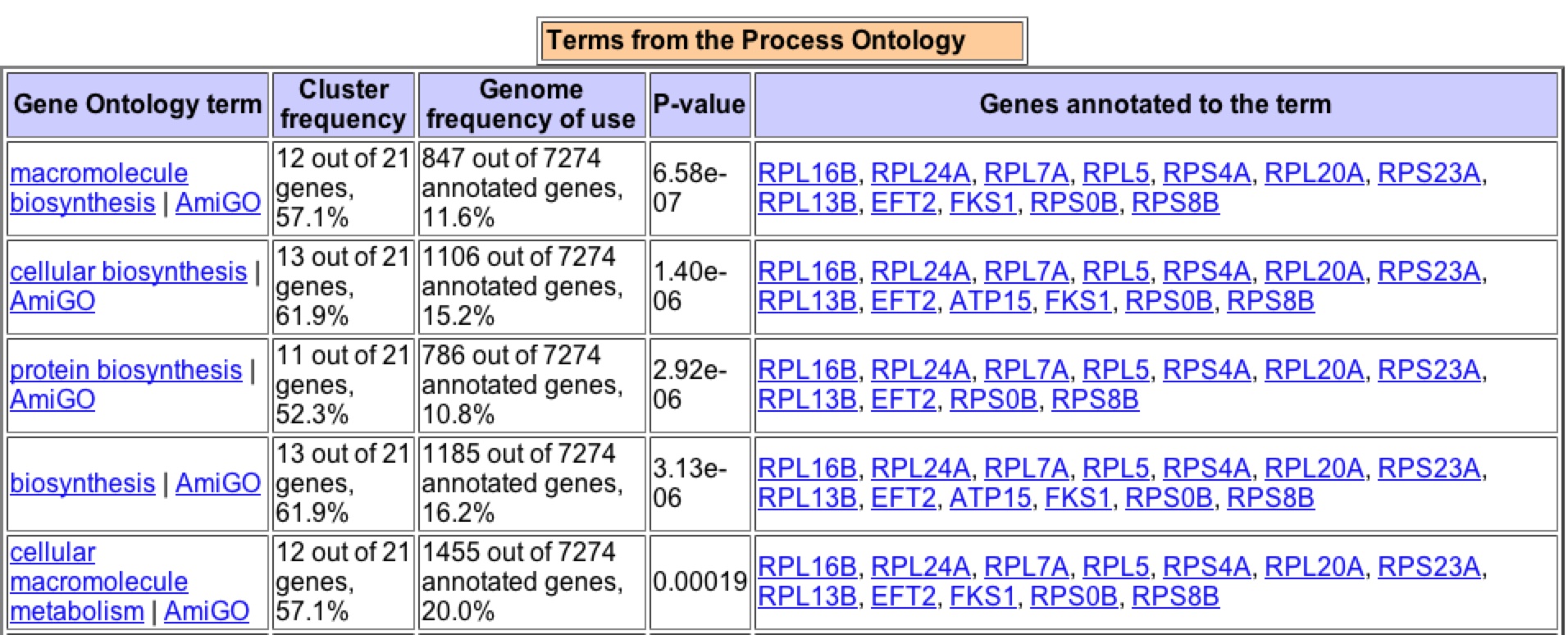

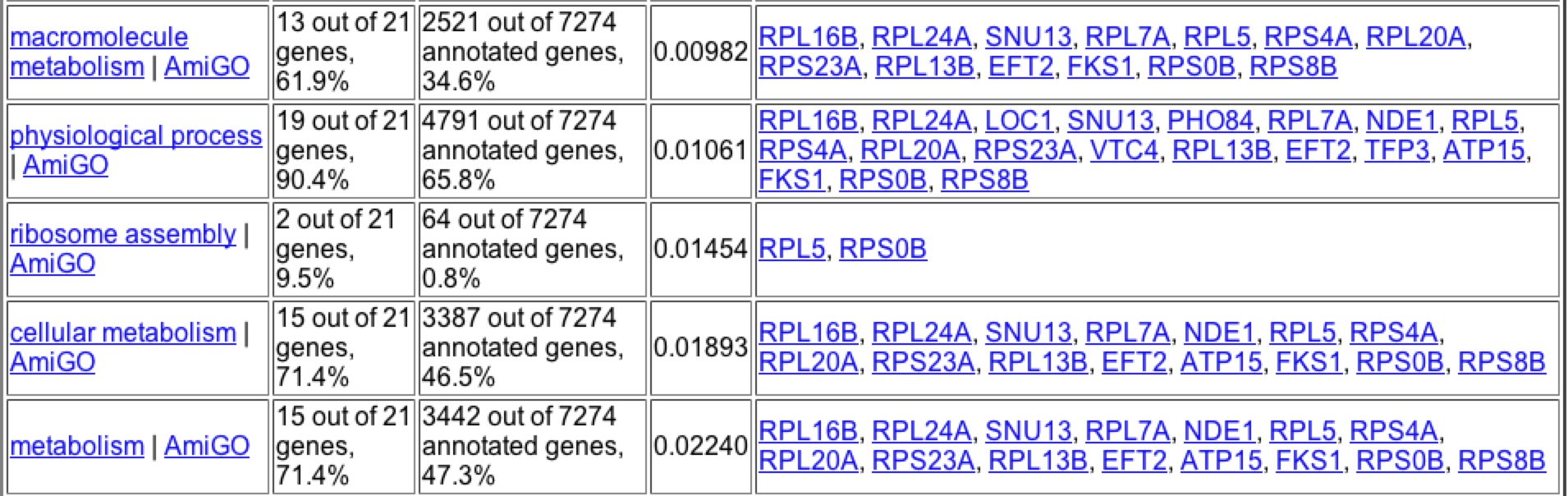
Figure 12: Expression patterns for YKL069W under histone depletion gave these significant Gene Ontology Terms for process. Image taken from the Gene Ontology Term Finder in Expression Connection.
Conclusions
Based on my findings regarding expression of YNK1, it seems very reasonable that it encodes a nucloside diphosphate kinase, as it is induced under DNA-damaging stress, and is probably under the control of the calcineurin transcription factor.
In regards to YKL069, I now believe that it is a non-membrane protein found near ribosomes that is involved in protein biosynthesis, and it is highly repressed during histone depletion and induced during nitrogen depletion, suggesting it may be involved in nitrogen freeing or storing processes. It also appears to be involved in the physiological processes of the cell through Guilt-by-association.
References:
Amutha B, Pain D. 2003. Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochemical Journal 370:805-815.
DeRisi JL, Iyer VR, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278(5338):680-6
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11(12):4241-57
Gene Ontology Software Group. 2003. Gene Ontology Database. <http://www.godatabase.org/dev/database>. Accessed 2005 Oct 18.
Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, Brown PO, Cyert MS (2002) Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem 277(34):31079-88
Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402(6760):418-21
Any questions or concerns regarding this page should be directed to magemberling "at" davidson.edu.