This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Proteins
So far, I have investigated two yeast genes within Saccharomyces cerevisiae, GDA1, and the non-annotated gene, YEL043W. For general information about these two yeast genes, click here. For information about the expression of these two yeast genes, click here.
This page will examine the proteins encoded by these two genes. Many public databases are available to search for information about proteins, including shape, size, function, and protein-protein interactions. Within this page, I will discuss some of these characteristics of GDA1p and YEL043W, as well as hypothesize the function of the non-annotated gene YEL043W and design an experiment to test my hypothesis.
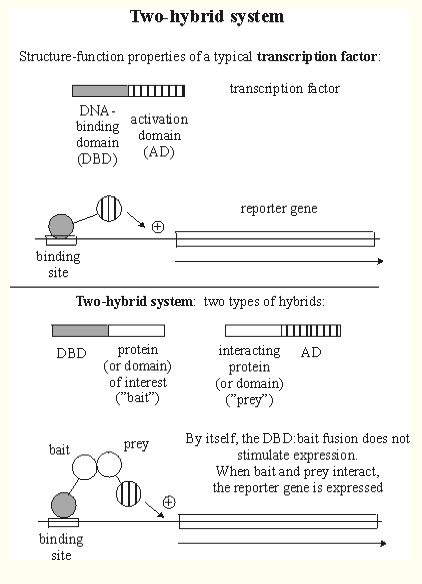
An important method for determining protein-protein interactions is yeast two-hybrid. In this method, a protein of interest, referred to as "bait," is bound to a DNA Binding Domain (DBD). A separate protein, called the "prey," is bound to an open reading frame. If these two proteins (the bait and prey) interact, a reporter gene is transcribed (Figure 1). For more information about the yeast two-hybrid method, click here.

Figure 1. This is a graphical explanation of two-hybrid protein interactions. The DNA-binding domain (DBD) hybrid binds to the "bait" (protein of interest), but cannot activate transcription. However, after binding with the activation domain (AD), the two genes interact, enabling the expression of the reporter gene. The protein of interest can be isolated using the reporter gene as a marker. Image from http://www.biochem.arizona.edu/classes/bioc568/two-hybrid_system.htm. Permission pending.
GDA1p
The Comprehensice Yeast Genome Database (CYGD) provides general information about proteins, including some basic physical features, such as size and location (Figure 2).

Figure 2. The above image provides information about GDA1p, including location, length, molecular weight, and transmembrane domains. Image courtesey of http://mips.gsf.de/genre/proj/yeast/. Permission pending.
CYGD also provides some insight into the function of GDA1p (Figures 3 and 4).

Figure 3. The remarks in the above image report that GDA1p has a signal sequence that is 29 amino acids long. It also reports on the location of the protein at various times of its cycle (travels between the lumen and cytoplasm). This information is important when analyzing the function and interactions of proteins. Image from http://mips.gsf.de/genre/proj/yeast/. Permission pending.

Figure 4. From CYGD we learn GDA1p is involved in metabolic pathways, specifically glycosylation. CYGD also provides a link to 69 studies discussing GDA1's role in glycosylation. MIPS also reports that GDA1p is involved in glycosylation, the process of adding a glycosyl group to a protein to create a glycoprotein. Image from http://mips.gsf.de/genre/proj/yeast/. Permission pending.
Protein Information Resource (PIR) is another useful database when researching the function and interactions of particular proteins. PIR provides useful research tools like accession numbers, multiple protein names, and the protein sequence (Figure 5).
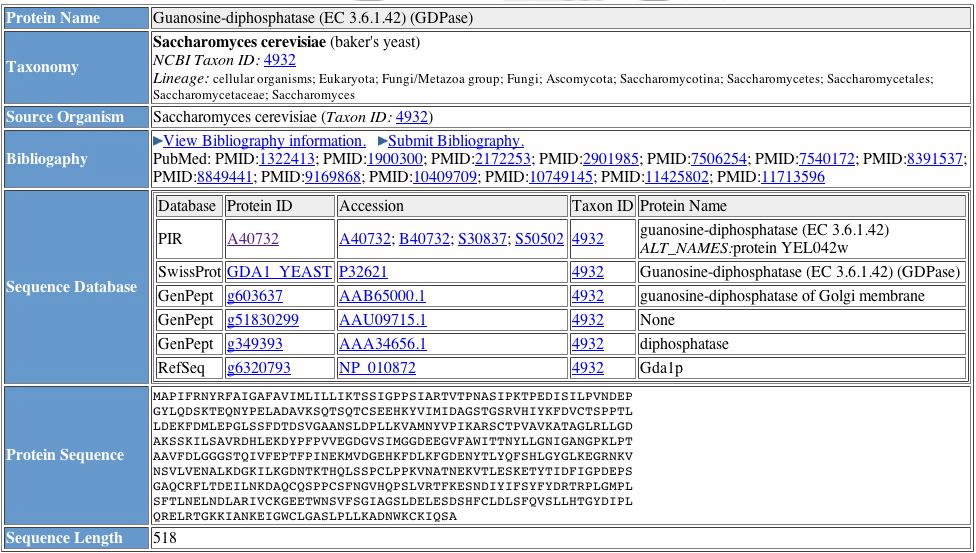
Figure 5. Results from a PIR protein query. Information such as protein name and accession number are extremely helpful when working with several databases. Image from http://pir.georgetown.edu/cgi-bin/nfEntry.pl?id=NF00366590. Permission pending.
| Figures 2 through 5 provide general information about GDA1p. From Figure 1, we learn GDA1p is 518 amino acids long and has one transmembrane domain, confirming findings mentioned on the My Favorite Yeast Genes page. Figures 3 and 4 both confirm the idea that GDA1p is involved in glycosylation, converting GDP to GMP. Figure 5 provides the protein sequence of of YEL043W and accession numbers which are important when keeping track of proteins when searching multiple databases. The four figures above compose a fairly comprehensive package of the general characteristics and function of GDA1p. |
The Database of Interacting Proteins (DIP) provides visual representations of protein-protein interactions. In figure 6, the large red dot represents GDA1p. The orange dots represent proteins only one edge away, and yellow dots represent proteins that are two edges away from GDA1p. Refer to the legend for further explanation of the diagram (Figure 7).
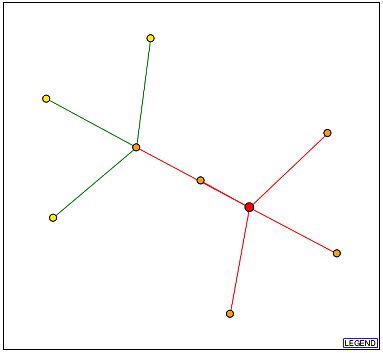
Figure 6. Above is a visual representation of the protein-protein interactions of GDA1p. The red dot represents GDA1p. Moving clockwise beginning with the dot above and to the right of GDA1p, the first shell nodes are Ald5p, Ssp120p, YEL017W, YJL152W, and Hpa2p. Figure from http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=5284. Permission pending.
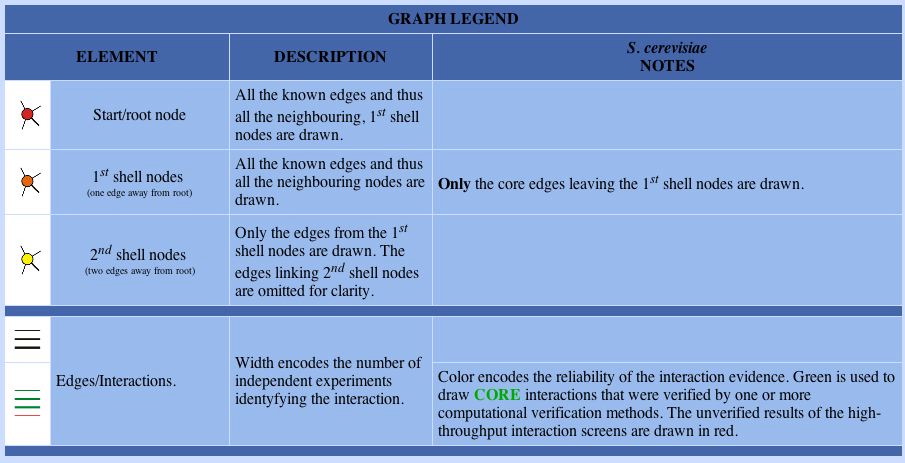
Figure 7. The legend for the DIP image. First shell proteins directly interact with the starting protein, GDA1p in this case, and second shell proteins interact with the first shell proteins. Image courtesy of http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=5284. Permission pending.
| Without Figure 7, Figure 6 does not provide too much information. Using the legend, we learn about GDA1p's protein-protein interactions. According to Figure 6, GDA1p engages in five direct interactions and three second shell interactions. The thin lines between all the nodes tell us few independent experiments have been conducted to confirm these interactions. However, the second shell interactions are colored green, indicating these interactios have been verified by multiple computational methods. |
Knowing the functions of the proteins with which GDA1p interacts allows from a more complete understanding of GDA1p itself. The Yeast Grid database lists proteins that interact with GDA1p, as well as their GO annotations and a description of the protein (Figure 8).
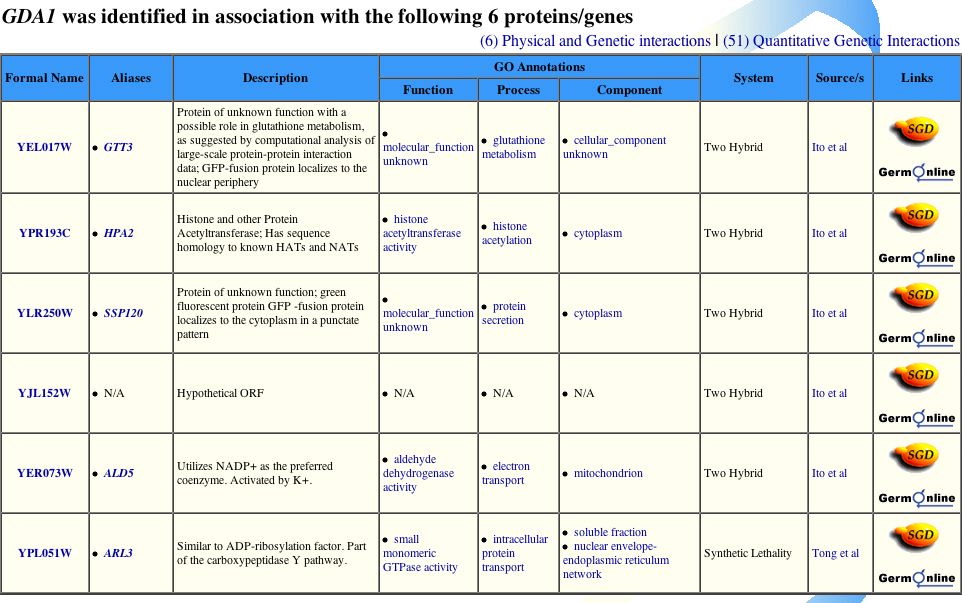
Figure 8. The proteins listed above match all but two of the proteins from the DIP image. All but one of the protein-protein interactions were discovered through the yeast two-hybrid method described above. Image from http://biodata.mshri.on.ca/yeast_grid/servlet/SearchResults. Permission pending.
Comparing the Yeast Grid protein interactions to the MIPS Protein-Protein Interaction database yields the same results as the DIP image (Figure 9).
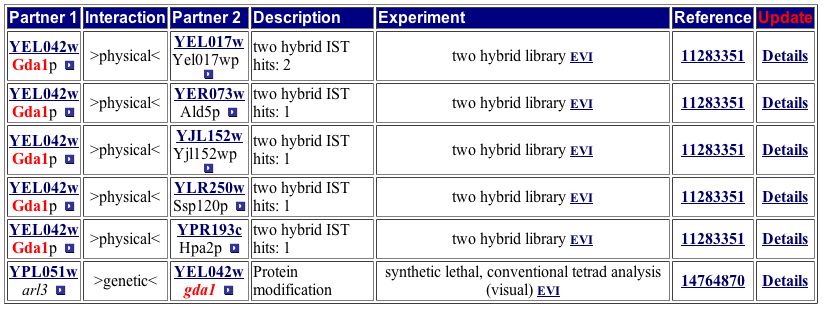
Figure 9. This table of GDA1p interactions matches the one from the Yeast Grid, neither of which directly match the image from DIP. Both of the tables include the genetic interaction of ARL3. Image from http://mips.gsf.de/proj/yeast/CYGD/interaction/ . Permission pending.
The above two tables are very important for understanding the details of GDA1p's function. The two tables include the same six protein interactions. Figure 8 also includes any known functions of the six other proteins. The proteins with which GDA1p interacts are located in the cytoplasm, ER, and mitochondria and participate in myriad pathways. Since many types of proteins interact with GDA1p, it probably fulfills a role vital to the performance of most proteins. The processes listed in Figure 8 are nearly all involved in some sort of protein modification, as is GDA1p. Both tables include ARL3 as a protein with which GDA1p interacts, but the DIP image does not include this protein. One possible explanation is that ARL3 was identified as part of a protein-protein interaction with GDA1p through synthetic lethality instead of yeast two-hybrid, causing it to be excluded from the DIP image. |
I searched the following databases for more information about GDA1p protein interactions, but found little to no relevant information:
Enzymes and Metabolic Pathways Database
YEL043W
Analysis of physical features and the protein interactions for YEL043W is crucial to determining this non-annotated gene's function. As explained in the web page addressing the expression of YEL043W, understanding the genes with which an unknown gene interacts, the Guilt by Association principle can be envoked to allow for grounded predictions to be made about the gene. The same sites as above were used to gather information about YEL043W.
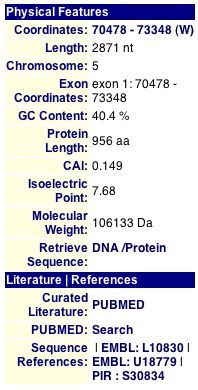
CYGD describes YEL043W as a "protein of unknown function localized to ER" (CYGD, 2005). This database also includes information on the protein's physical features (Figure 10). Unfortunately, no YEL043W homologs exist according to CYGD, making an analysis of the function of YEL043W a bit more difficult. Compared to the annotated GDA1, YEL043W has much less information within CYGD.
![]()
Figure 10. The above image provides information about YEL043W, including location, length, molecular weight, and transmembrane domains. CYGD also has no remarks for the non-annotated protein YEL043W. Image courtesey of http://mips.gsf.de/genre/proj/yeast/. Permission pending.
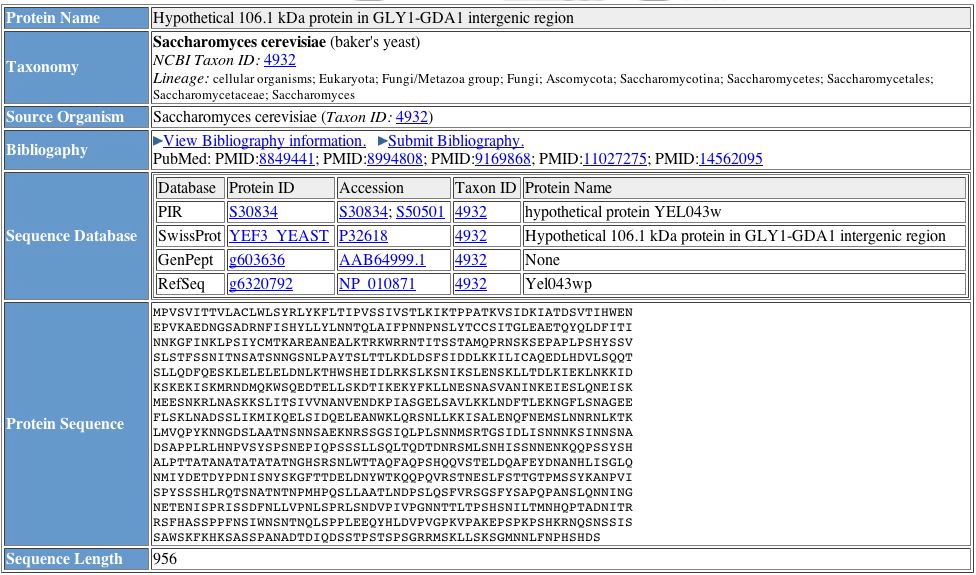
Figure 11. General information about YEL043W, including the protein's amino acid sequence. The listed databases contain limited information about YEL043W. Image courtesy of http://pir.georgetown.edu/cgi-bin/nfEntry.pl?id=NF00363897. Permission pending.
| Figures 10 and 11 provide general information about YEL043W. We learn YEL043W is 956 amino acids long, and Figure 11 provides the actual sequence. We also discover YEL043W's target protein is a secretory pathway of 29 amino acids. In Figure 11, SwissProt records YEL043W as a hypothetical protein within the GLY1-GDA1 intergenic region, but no function is mentioned in either of these figures. |
A DIP query yields an image more complex than GDA1p's DIP image (Figure 12). This seems somewhat odd considering the lack of information available regarding YEL043W.
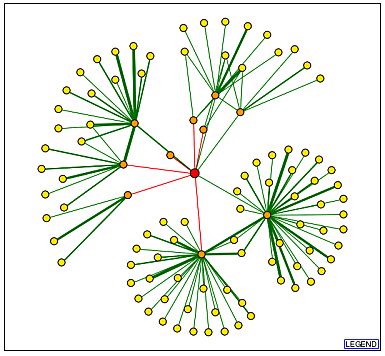
Figure 12. YEL043W has many more first and second shell shell nodes than GDA1p. Image from http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=2682. Permission pending.
| Using information provided by the legend from the previous DIP image, we learn YEL043W has 10 first shell nodes, and many more second shell nodes. Some of the second shell nodes have thick, green lines, indicating independent verification of the particular protein-protein interaction. According to DIP, YEL043W has many more protein-protein interactions than GDA1p. This surprises me since little is known about the function of YEL043W. It could be possible, therefore, that these are hypothetical interactions, and a portion of them do not occur, but cannot yet be ruled out. It is also possible that all of these interactions do in fact occur, but the details have yet to be learned. |
Yeast Grid does list six proteins with which YEL043W interacts (Figure 13).
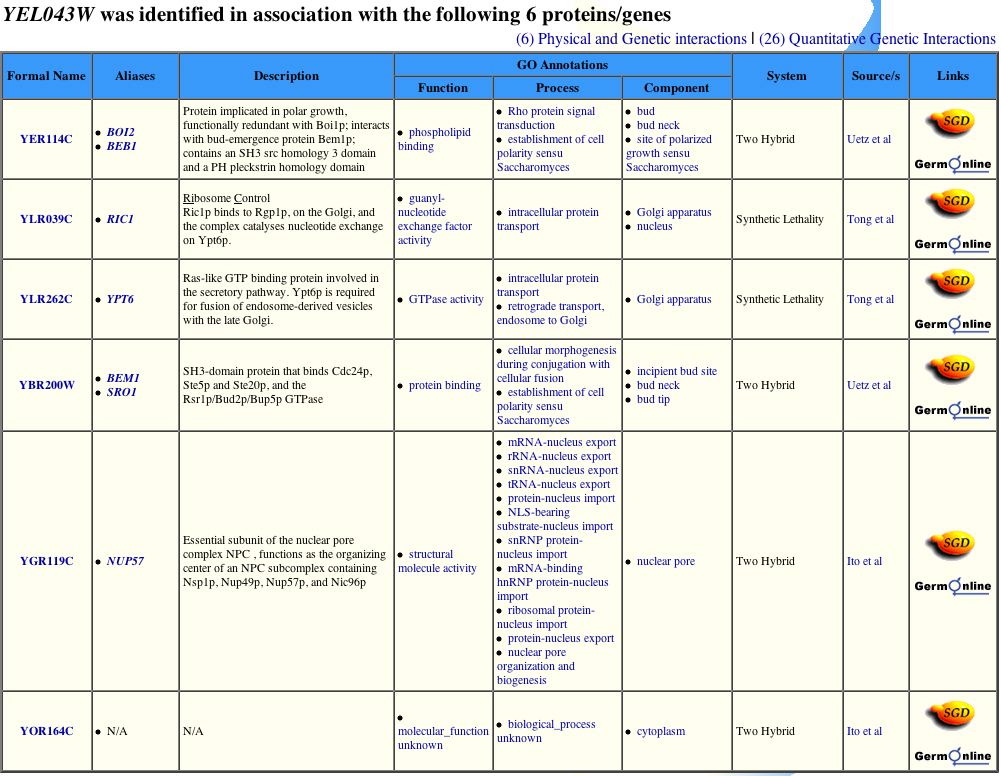
Figure 13. Yeast Grid provides a list of six proteins with which YEL043W interacts. However, according to the DIP image, YEL043W interacts with 10 first node proteins. Image from http://biodata.mshri.on.ca/yeast_grid/servlet/SearchResults. Permission pending.
CYGD provides a more extensive list of protein-protein interactions, returning 11 hits (Figure 14). Some of these hits match the DIP image while others do not.
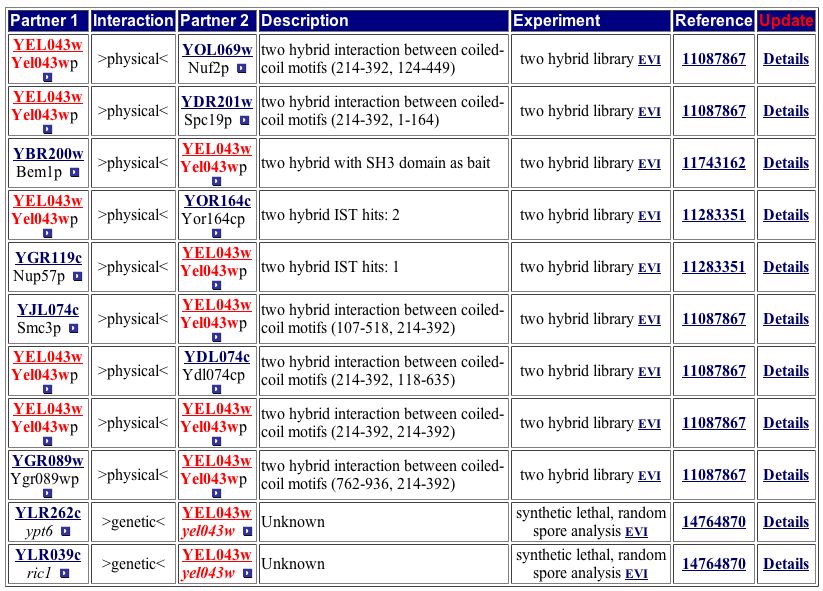
Figure 14. The CYPD has one more protein listed than Yeast Grid. Two of the 11 proteins are listed as genetic interactions. Image from http://mips.gsf.de/proj/yeast/CYGD/interaction/. Permission pending.
The YRC two-hybrid analysis database provides a list of protein-protein interactions within S. cerevisiae. Below are two listings that include YEL043W (Figure 15).
![]()
![]()
Figure 15. The above image outlines two protein-protein interactions involving YEL043W observed through the yeast two-hybrid method. As the headings state, the protein in the left most column acts as the bait (attached to the DNA binding domain), and the middle column is the prey protein (fused to the activation domain), and the prey ORF is the gene transcribed if the bait and prey interact. Image from http://depts.washington.edu/%7Eyeastrc/th_11.htm. Permission pending.
| Only three protein-protein interactions are consistent between DIP and Yeast Grid. This discrepency could represent incomplete analysis of currently available data. Only four protein-protein interactions are consistent between DIP and the CYPD table (Figure 14). Yeast Grid and CYPD include two protein-protein interactions (RIC1, YPT6) absent from the DIP image. Although BEM1 appears in all four sources, including the YRC two-hybrid analysis database, BOI2 is missing from the CYGD table. The proteins common across at least three of the four databases are either involved in establishing cell polarity or protein transport. The locations of these proteins switch between the Golgi, nucleus, and cytoplasm. The discrepencies between these databases indicate more research is needed to build a clearer profile of YEL043W, but trends from the current data suggest YEL043W is invovled in the transport of proteins. I therefore change my hypothesis from YEL043W aiding in the maintenance of cytoskeleton architecture to a gene that encodes a protein involved in protein transport from ER to the Golgi. |
The following databases had no new information regarding YEL043W:
Enzymes and Metabolic Pathways Database
Theoretical Experiment to learn more about YEL043W
So far, we have gathered data to support the hypothesis that the protein encoded by YEL043W aids in protein transport. According to DIP and PIR, YEL043W interacts with a multitude of proteins located in the nucleus, cytoplasm, and ER, and CYGD reports that YEL043W is located in the lumen of the cell. This fact suggests YEL043W aids in transporting proteins from the ER to the Golgi, where they then undergo modification and are taken to their primary work place.
To determine whether this hypothesis is correct, I would first use immunofluorescence to determine the exact location of the protein. To do this, I would insert an epitope tag onto the end of a cloned YEL043W protein. I would then administer a primary antibody (hemagglutinin monoclonal antibody) to cells with the tagged protein. A second antibody is then necessary to bind to the first to create fluorescing. Under a fluorescent microscope, I could pinpoint the location of YEL043W to verify whether the protein actually stayed within the lumen, acts as an intergral membrane protein, or resides in another organelle involved in glycosylation, such as the Golgi.
Determining the protein's location allows me to then better understand probable protein-protein interactions. Because the portein-protein interaction data for YEL043W seems inconsistent, more yeast two-hybrid experiments need to be done. I would start by examining proteins within the lumen and proteins that might aid in actual protein transport from the ER to the Golgi. These proteins would serve as the prey, and YEL043W would serve as the bait in the yeast two-hybrid experiments. This would hopefully narrow down the list of proteins with which YEL043W interacts. I would follow up the experiment by only using portions of the YEL043W ORF as the bait to then learn where exactly the proteins interact with one another.
References
Comprehensice Yeast Genome Database (CYGD). 2005. <http://mips.gsf.de/genre/proj/yeast/>. Accessed 2005 Nov 16.
Database of Interacting Proteins (DIP). 2005. <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>. Accessed 2005 Nov 16.
Little, J. 2004. Two-hybrid system for predicting protein-protein interactions. <http://www.biochem.arizona.edu/classes/bioc568/two-hybrid_system.htm>. Accessed 2005 Nov 17.
Protein Information Resource (PIR). 2004. <http://pir.georgetown.edu/home.shtml>. Accessed 2005 Nov 16.
SGD (Saccharomyces Genome Database). 2005. < http://www.yeastgenome.org/ >. Accessed 2005 Nov 16.
Sobhanifar, Solmaz. The yeast two-hybrid assay: An excercise in experimental eloquence. <http://www.bioteach.ubc.ca/MolecularBiology/AYeastTwoHybridAssay/>. Accessed 2005 Nov 17.
Yeast Grid. 2005. <http://biodata.mshri.on.ca/yeast_grid/servlet/SearchPage>. Accessed 2005 Nov 16.
YRC. 2005. <http://depts.washington.edu/%7Eyeastrc/th_11.htm>. Accessed 2005 Nov 16.
Questions or comments? E-mail Caitlin Kiley at cakiley@davidson.edu
Return to Davidson College Biology Home Page
Return to Genomics Home Page
Return to Caitlin Kiley's Home Page
Return to Davidson College Home Page