This web page was produced as an assignment for an undergraduate course at Davidson College.
Cancer Genetics in the News
Introduction:
Popular press science writers have the difficult job of explaining scientific research, which involves specialized knowledge and a specialized language to go with it, in such a way that the general reader can understand the science and the issues surrounding it. Success in conveying this knowledge is not an easy feat.
As described by Kua et al. (2004), popular press science writers have three roles: to be an intermediary, a watchdog, and a tool-giver. The intermediary is the most obvious and in many ways the most important role for the writer. He or she must essentially translate the language of science into everyday English (or whatever language they are writing in).
The second role, a watchdog, is important for all journalism and calls the writer to discuss the implications, both social and ethical, of the research.
The role of tool-giver is to give readers the tools they need to evaluate the science and research. It involves describing the science and longer-term significance of the research, but it also involves giving a context of the work: explaining the knowledge of the field and the future applications of the research. This gives readers a context so that he or she can decide if the findings actually are huge news or just a step in an interesting direction.
Nature paper:
In June 2012, Nature published a paper entitled “Whole genome analysis informs breast cancer response to aromatase inhibition” by Ellis et al. about genes that play a part in breast cancer and how they respond to a certain type of treatment. Shortly after this, the Economist published an article in their print magazine stating that certain mutations can help predict which cancers will respond to therapy. The Nature paper describes a genome analysis of DNA from breast cancer tumors, sampled from people involved in a neoadjuvant aromatase inhibitor therapy, a possible treatment for the cancer. It describes the specific mutation that were found and how some of these mutations relate to how the person with the tumor responds to the aromatase inhibitor therapy.
Article in The Economist:
The review article in The Economist begins by placing the study in the wider framework of cancer research by describing the work of the International Cancer Genome Consortium, a group of laboratories around the world that are building a list of genetic mutations involved in cancer. It uses the example of breast tumors to show the quantity of data being produced: 3,000 breast tumors have been sequenced and published, but then goes on to say that these data in itself with not help patients until they are placed in the context of drug trials. The article then goes on the say that this is what the research team at Washington University in St Louis have done by sequencing tumors from patients involved in aromatase inhibitor therapy.
This article then gives an overview of the methods used by the researchers: to sequence DNA from both healthy tissue and cancerous tissue from the patients to determine what genes had mutated. They chose to carry out this study with this particular group of patients involved in the drug therapy because not all of the patients responded equally to the treatment.
The article then describes the results: the researchers found 18 frequently mutated genes, some of which had previously been found to be related to cancer genetics, as well as some that had not or had been connected to other forms of cancer, like leukemia. Specifically though they found that the patients whose tumors had a mutation in the p53 gene (Figure 1) less frequently respond favorably to the drug therapy, but those with mutation in the MAP3K1 or MAP2K4 genes were more likely to respond favorably to the drug.
The article concludes by bringing the study back out to the clinical level by describing the implications it has for treatment specifically the possibility of linking certain genetic mutations to the best drug to treat it.
Fulfilling the three roles:
The writer of this article seems to do a good job of filling the roles of intermediary and tool-giver as described by Kua et al. (2004), but it is harder to see how the writer fulfills the role of watchdog. First as an intermediary, the article translates well the technical terms used in the Nature article. It simplifies the methods to an understandable level, while still explaining what was done. It also describes in a clear and concise way the general understanding of the genes: some had already been linked to cancer, some were linked with leukemia, p53 regulates DNA repair, cell division and cell suicide (and therefore mutations to it can lead to cancer), and the MAP genes promote cell growth.
The fulfillment of the role of watchdog is harder to see. They do describe the social implications of the research in that it could revolutionize how cancer is treated, and yet also are cautious to say that it will soon revolutionize treatment. This role is not as completely filled because the writer does not discuss any ethical implications.
The writer of this article definitely fulfills the role of a tool-giver by contextualizing the research in the broader field of cancer research. The writer begins with explaining the International Cancer Genome Consortium and the large quantities of data they are coalescing and then narrows in to show how this research helps to push the knowledge to a new level by connecting it with drug therapies. The writer then ends with how the research could lead to new ways to treat cancers, even beyond breast cancer. However, though the writer explains how future treatment could work, he or she is careful to say that the research has implications for treatment, not certainties, and the research has not gone so far as to know what drugs to use. The information that is given and the understandable way the research is described give the reader a context within cancer and genomics research to understand how this study moves cancer genomics research towards treatment.
This article is also a good piece of pop science writing because, in addition to being understandable, it contains all the parts of a normal science paper: an introduction, description of methods, presentation of results, and discussion of conclusions and implications. The fact that it contains a description, if very simple, of the methods used and the way that they improve upon previous methods, shows an understanding of the way science works.

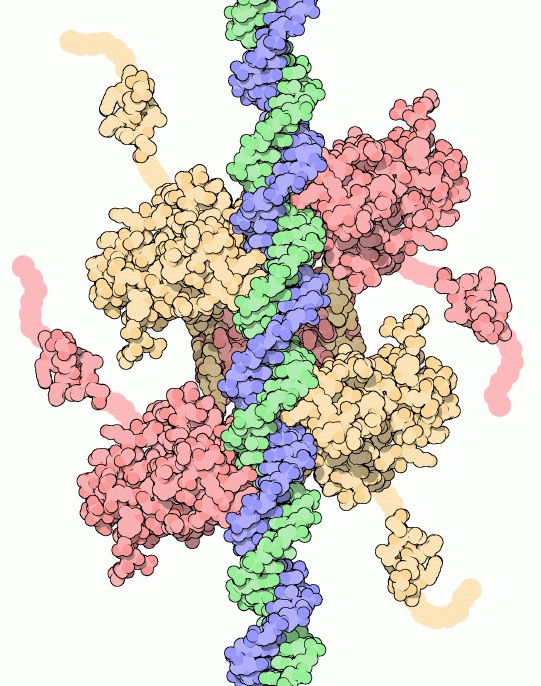
Figure 1. This shows the p53 protein (called the “p53 tumor suppressor” by the Protein Data Bank) hugging DNA with all four of its arms. This protein suppresses tumor formation by attaching itself to DNA to repair the DNA. When the gene encoding this protein was mutated in the breast tumor genome, the patients did not as frequently respond favorably to the aromatase inhibition therapy. Picture courtesy of the Protein Data Bank.
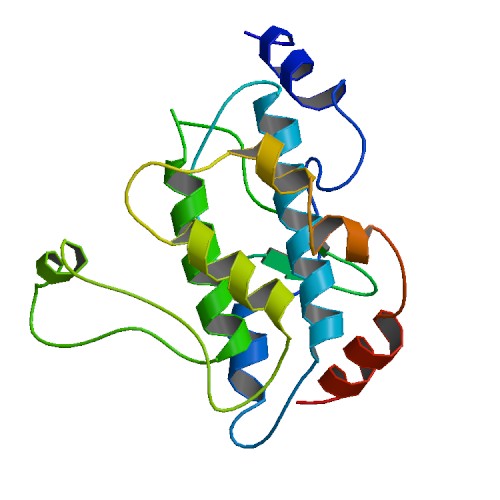
Figure 2. This is a structure of the non-phosphorylated MAP2K4 protein. When the gene encoding this protein was mutated in the breast tumor genome, the patients more frequently responded favorably to the aromatase inhibition therapy. Picture courtesy of the Protein Data Bank.
References:
Ellis, Matthew, et al. 2012. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012;486: 353-360.
Kua, Eunice, Michael Reder, and Martha J. Grossel. Science in the News: A Study of Reporting Genomics. Public Understanding of Science 2004;13: 309–322.
Gene Therapy: Genetic mutations predict which cancers will respond to treatment. Economist 2012;June 16 (From print edition)
Genomics Page
Biology Home Page
Email Questions or Comments to Becca Evans.
© Copyright 2013 Department of Biology, Davidson College, Davidson, NC 28035