This web page was produced as an assignment for an undergraduate course at Davidson College
Strengths and Weaknesses of Genomics Reporting in the Popular Press: Describing Dog Domestication
Which of these two articles are you more likely to read: “Learning to love grains, potatoes was the key to the evolution of dogs” or “The genomic signature of dog domestication reveals adaptation to a starch-rich diet?” I would argue that most people choose the former. Though both articles present the same discovery, the first title tells a better story and does so in language that anyone can understand.
Science reporting is an integral step in the dissemination of research knowledge. The general public is not only unlikely to access articles in peer-reviewed journals, but also often unable to understand the field-specific jargon that these papers employ. Genomics papers, for instance, use specialized vocabulary to describe many things including the gene(s) or protein(s) studied, the data collection methods, and the data analysis methods. Nonetheless, genomics research often produces findings that are either important or interesting to a wide audience. Going back to the two articles mentioned above, lots of people love dogs and would be interested to know how they became the way they are today.
However, several major criticisms have been leveled against popular press science reporting. In “Science and the news: A study of reporting genomics” Kua et al. (2004) conclude that the popular press often differs in “what is said rather than how it is said.” While they fully acknowledge the limitations that reporters face in terms of space and scientific literacy, Kua et al. argue that science reporters need to focus more on the methods used and to give readers a better sense of the findings’ context (i.e. to discuss the current body of knowledge on the topic). Ideally, science reporting should provide sufficient detail to give readers the “tools” to think for themselves (Kua et al., 2004). The following popular press “translation” of genomics research provides one example of a fairly successful application of the recommendations made by Kua et al.
Original Research Findings: Axelsson et al., 2013
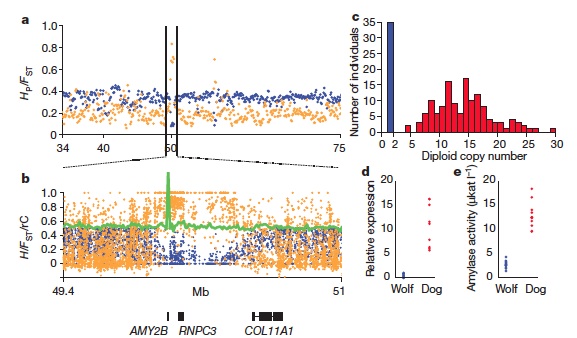
The article “The genomic signature of dog domestication reveals adaptation to a starch-rich diet,” published in Nature provides insight into the genetic differences between dogs and wolves. The authors used whole genome sequencing to identify genomic regions that may have been selected for during domestication (regions with reduced heterozygosity and increased genetic distance from the wolf genome). A number of the regions they identify contain genes related to brain function, but the authors conduct more detailed analysis of amylase, maltase, and intestinal glucose transporter genes, all of which are more prevalent in dogs than wolves and confer an increased ability to digest starches. (See Figure 1 for a visual representation of the authors’ amylase analysis, analysis process was similar for other genes). The main point conveyed in the article is that starch digestion adaptations allowed modern dogs to diverge from wolves and were an important step in the domestication process.

Figure 1. (a) and (b) In the amylase gene coding region a plot of pooled heterozygosity (blue) indicates little variation among dogs for this coding region; a plot of average fixation index (orange) indicates high divergence from the wolf genome for this coding region. The green peak indicates a high relative copy number of AMY2B genes in dogs. (c) A plot of the copy numbers for AMY2B genes in dogs (red) and wolves (blue); an increased number of diploid copies in dogs allows the increased mRNA expression for the AMY2B genes seen in (d) and increased amylase activity seen in (e). Image reprinted by permission from Macmillan Publishers Ltd: Nature (Axelsson et al., 2013).
Popular Press Report: Brown, 2013
The article “Learning to love grains, potatoes was the key to the evolution of dogs,” published in the Washington Post conveys many of the details of the study well, but overstates its implications in terms of the way dogs became domesticated. The author gives his readers many of those details about context and methodology that Kua et al. find lacking in the popular press. Brown goes into great detail about the debate that surrounds dog evolution and domestication. One theory proposes that dogs began scavenging near human settlements and eventually “domesticated themselves” to cohabitate better with humans, the other suggests that humans captured wolves an then bred them for guarding or hunting. Brown also provides a clear, if simplistic, explanation of the researchers’ methodology stating that the team “compared the genomes of wolves and dogs… and identified 36 genomic regions in which there are differences that suggest they have undergone recent natural selection in dogs.” Brown goes on to identify the three starch-digestion genes that the team examined in greater detail. He even explains the importance of the many amylase gene copies seen expressed in dogs. Overall, I think it is fair to say that this article provides sufficient contextual and methodological detail to satisfy the standards set out by Kua et al.
Nonetheless, this article does seem to justify the complaint that popular press reporting differs in “what is said rather than how it is said” (Kua et al., 2004). Though the original research does touch on the dog domestication debate, and seems to provide conditional support for the scavenger hypothesis, this argument becomes central to the Washington Post article. The original researchers devote a total of about four sentences to outlining the two hypotheses and stating that their work “may suggest that a change of ecological niche could have been the driving force behind the domestication process, and that scavenging in waste dumps…may have constituted this new niche” (Axelsson et al., 2013). By contrast, Brown uses the researcher’s findings mainly to justify the “self-domestication” hypothesis and appeals to other dog evolution experts to lend further support to the argument. Even as Brown’s argument strays from the main thesis of the study, however, he does not overstate the researcher’s findings saying that, “evidence of natural selection in the number and efficiency of key digestive enzymes supports the hypothesis that dogs may have domesticated themselves as a way to exploit the garbage of permanent human settlements” (Brown, 2013).
Overall, Brown’s “translation” of the Axelsson article is successful because it keeps the general public well informed about the scientific facts, context, and methodology of this study. Though the Washington Post article goes beyond paraphrase and presents a more contentious argument than the original authors, it does not misrepresent the authors’ findings in doing so.
Works Cited