
This web page was produced as an assignment for an undergraduate course at Davidson College.
Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Gliomas: A Summary

From Johnson et al., 2014
Johnson et al. used genomics to answer two questions with regard to recurrent human gliomas: 1) “What is the extent to which mutations in initial tumors differ from their subsequent recurrent tumors?” And 2) “How does chemotherapy with temozolomide (TMZ), a drug commonly used in the treatment of glioma, affect the mutational profile of recurrent tumors?” (Johnson et al., 2014). In other words, do recurrent tumors have different mutations than the initial tumors, and does TMZ treatment affect the rate at which new mutations occur?
Using comparative exome sequencing of the initial tumors and recurrent tumors after TMZ treatment and taking into account intratumoral heterogeneity (the possibility that “geographically distinct parts of the tumor may have different mutations”), the researchers found that not only do many recurrent tumors have different mutations than the initial tumors, TMZ treatment often induces driver mutations that result in more aggressive glioblastoma (GBM) (Johnson et al., 2014). These findings have implications for tumor treatment and future genomic research aimed at developing therapies. In addition, they traced the phylogeny of the mutations to determine when new mutations that led to recurrences occurred.
Overall, the research outlined in this paper is well controlled and conducted. However, I would like to see sequencing of more than just the exomes. With full genome sequencing readily available, sequencing introns as well as exons allows researchers to validate their sequencing and provides wild-type data for cancerous cells. While the researchers did analyze portions of the transcriptome, whole genome sequencing (and transcriptome sequencing too) would also provide evidence for mutations that lead to alternate splicing. The data presented here is very convincing, but some of the figures could be improved by choosing more contrasting colors (ex. Figure 3D). Additionally, while the phylogenetic trees in Figure 2 are convincing evidence that the difference in the initial and recurrent tumors is due mainly to evolution over time and not to intratumoral heterogeneity, the pictures of the tumors do little to improve the reader’s understanding of the data. Finally, the researchers’ argument against intratumoral heterogeneity as the primary explanation for differences in initial and recurrent tumors is very convincing and well supported mainly because of their identification and quantification of TMZ-associated mutations in the recurrent tumors. It would, however, be nice to see how frequently these same mutations occur in tumors that are not exposed to TMZ.
Figure 1A-C addresses the first question, do recurrent tumors have different mutations than the initial tumor. Using 125-fold coverage exome sequencing, the researchers concluded that many recurrent tumors have a large number of novel mutations. Figure 1D looks specifically at the effect of intratumoral heterogeneity in order to support their claim that recurrent tumors do in fact have novel mutations and their data is not just an artifact of where in the tumor they were sequencing. Using exome sequencing and droplet digital polymerase chain reaction, they showed that intratumoral heterogeneity could pose a serious threat to the integrity of their data.
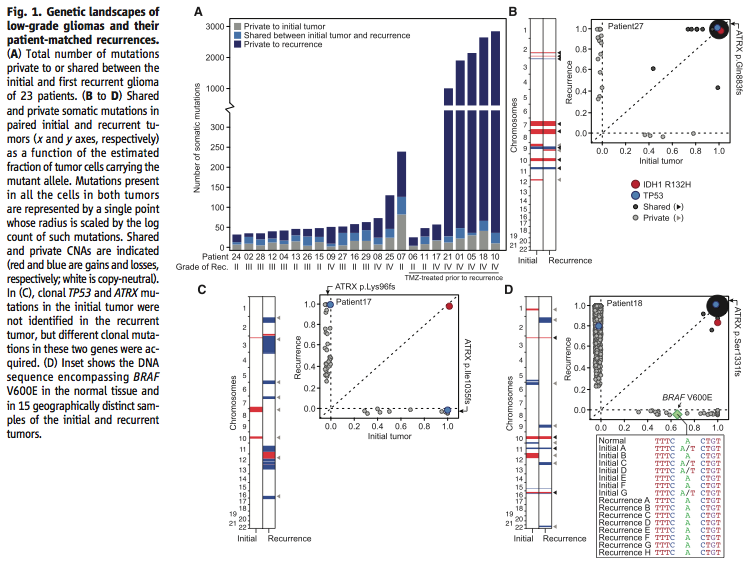
Figure 2 further addresses the question of the effect of intratumoral heterogeneity on the reliability of the data. Using exome sequencing, the researchers showed that although there is intratumoral heterogeneity, the initial and recurrent tumors “were only distantly related and … exonic mutations other than IDH1 R132H were only transiently present during the course of this patient’s disease” (Johnson et al., 2014). In other words, while both the initial and recurrent tumors have a certain amount of intratumoral heterogeneity, they are related phylogenetically and share only “a small minority of private mutations”, which means that intratumoral heterogeneity cannot account for most of the differences between the two tumors (Johnson et al., 2014).
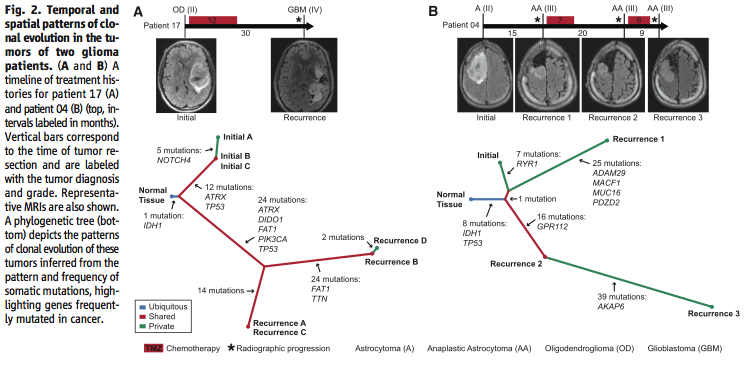
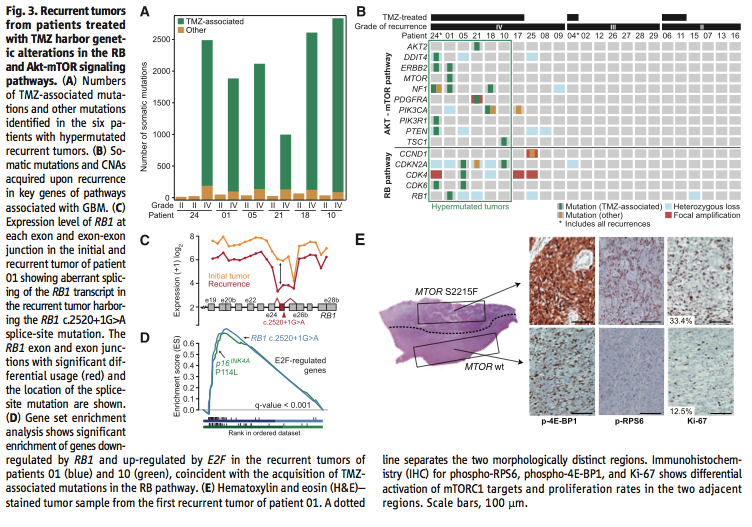
Figure 3 addresses the second big question, does TMZ treatment affect the rate at which new mutations occur in recurrent tumors. Using exome sequencing once again in Figure 3A-B to identify mutations commonly associated with TMZ exposure, and taking into account intratumoral heterogeneity, the researchers show that recurrences after TMZ treatment accumulate many TMZ associated mutations that lead to a “high-grade tumor with a worse prognosis” (Johnson et al., 2014). Using transcriptome sequencing in Figure 3C they confirm that a particular mutation TMZ-associated splice-site mutation “triggered aberrant splicing, premature termination, and loss of the RB1 C-terminal domain necessary for growth suppression”, which could lead to a more aggressive tumor (Johnson et al., 2014). The researchers used gene set enrichment analysis in Figure 3D to confirm that the mutation in Figure 3C occurred more often in recurrent tumors than initial tumors (i.e. was likely caused by TMZ treatment), and that this mutation did in fact “compromise the function of the RB tumor suppressor pathway” (Johnson et al., 2014). Finally in Figure 3E the researchers show through microdissection, immunohistochemistry, and hematoxylin and eosin staining that “TMZ-associated mutations conferred a proliferative advantage” and were responsible for starting the recurrent tumors (Johnson et al., 2014).
Sources:
Johnson, Brett E. et al. "Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma." Science 343:6167 (2014): 189-193. Web. 20 February 2014. Available http://www.sciencemag.org/content/343/6167/189.abstract
Assignments:
Assignment #3: You get to choose (due Apr. 25, 2 pm)
Genomics Page
Biology Home Page
© Copyright 2014 Department of Biology, Davidson College, Davidson, NC 28035