Overview:
This article examines the evolution of human diversity in specific geographical regions of our globe using The Simons Genome Diversity Project (SGDP). SGDP provides a more in-depth analysis of human genomes as they examine populations that were not particularly assessed in previous studies of this type. Their findings supported previous studies in that sub-Saharan populations had the highest genetic diversity. However, SGDP led to some novel results that were not seen in previous studies and perhaps could be explained by environmental and behavioral shifts affecting the human race. They found that East Asians have the highest proportion of Neanderthal ancestry of any population studied, and that there is greater Denisovan ancestry in areas of South Asia than other Eurasian populations. SGDP also provides some insight onto potential time frames of ancient population separations and provides possible explanations as to the what might support those claims. The authors are confident that SGDP can lead to more accurate discoveries about human evolution by analyzing rates of genetic variation among human populations.
Opinion:
I appreciate how the authors are using SGDP to try to develop a more accurate understanding of human genome evolution and migration by including a large amount of populations (142) in their studies. However, one specific part of this article that threw me off and made me lose some faith in their results was on page 204 when the authors stated "we caution that the date estimates also do not take into account uncertainty about the true value of human mutation rate, which could plausibly be 30% higher or lower than the point estimate we use" (Mallick et al. 2016). This means that their estimates on certain ancient human population divergences could very well be inaccurate because there is so much uncertainty in the value of the human mutation rate. They claimed that genome variation in divergent sites per base pair can be used to reconstruct population size changes and separations, and then they provide all of their estimates about time periods for multiple population divergences. That section is then wrapped up by their truthful statement that the provided dates must be taken with a grain of salt because of the amount of uncertainty in human mutation rate. The authors are honest about what could possibly make their results not accurate, and I would definitely rather have that than authors who attempt to mask their imprecision in their data. Overall, I believe this article provides a great explanation of SGDP, how it is different from previous genome projects, and the limiting factors that prevent the results from being completely accurate. This study leads to results that point towards a possible new train of thought on human evolution and migration history, and new opinions are essential to driving science in the right direction.
This article examines the evolution of human diversity in specific geographical regions of our globe using The Simons Genome Diversity Project (SGDP). SGDP provides a more in-depth analysis of human genomes as they examine populations that were not particularly assessed in previous studies of this type. Their findings supported previous studies in that sub-Saharan populations had the highest genetic diversity. However, SGDP led to some novel results that were not seen in previous studies and perhaps could be explained by environmental and behavioral shifts affecting the human race. They found that East Asians have the highest proportion of Neanderthal ancestry of any population studied, and that there is greater Denisovan ancestry in areas of South Asia than other Eurasian populations. SGDP also provides some insight onto potential time frames of ancient population separations and provides possible explanations as to the what might support those claims. The authors are confident that SGDP can lead to more accurate discoveries about human evolution by analyzing rates of genetic variation among human populations.
Opinion:
I appreciate how the authors are using SGDP to try to develop a more accurate understanding of human genome evolution and migration by including a large amount of populations (142) in their studies. However, one specific part of this article that threw me off and made me lose some faith in their results was on page 204 when the authors stated "we caution that the date estimates also do not take into account uncertainty about the true value of human mutation rate, which could plausibly be 30% higher or lower than the point estimate we use" (Mallick et al. 2016). This means that their estimates on certain ancient human population divergences could very well be inaccurate because there is so much uncertainty in the value of the human mutation rate. They claimed that genome variation in divergent sites per base pair can be used to reconstruct population size changes and separations, and then they provide all of their estimates about time periods for multiple population divergences. That section is then wrapped up by their truthful statement that the provided dates must be taken with a grain of salt because of the amount of uncertainty in human mutation rate. The authors are honest about what could possibly make their results not accurate, and I would definitely rather have that than authors who attempt to mask their imprecision in their data. Overall, I believe this article provides a great explanation of SGDP, how it is different from previous genome projects, and the limiting factors that prevent the results from being completely accurate. This study leads to results that point towards a possible new train of thought on human evolution and migration history, and new opinions are essential to driving science in the right direction.

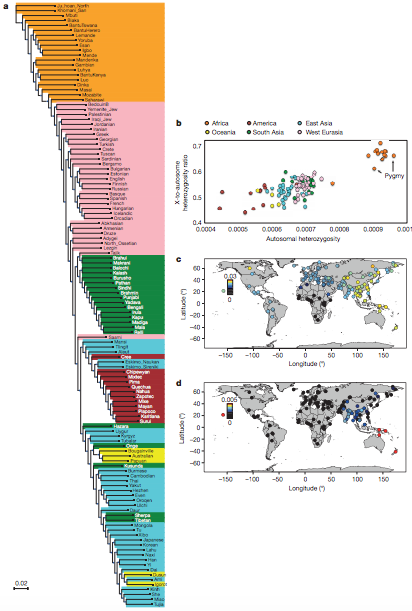
Figure 1: Genetic
variation in the SGDP. (Malllick et al. 2016)
Figure 1A: By analyzing pairwise divergence per nucleotide in the data obtained from SGDP, the authors were able to construct a large neighbour-joining tree. These results agree with previous studies in that the deepest splits in human population evolution are between the African populations. The African populations are in the orange at the top of the tree and their branches split the farthest to the left, indicating that their populations diverged before any of the others that are included. Before forming this tree, the authors carried out studies on population relationships with ADMIXTURE and principle component analysis which both form inter-population relatedness reads using differences in genetic variability.
Figure 1B: This panel examines the heterozygosity of modern human populations from across the world by comparing the proportion of diallelic genotypes per base pair between populations.The data agrees with previous studies in that sub-Saharan African populations and pygymies (orange dots) have both the highest genetic diversity and ratio of X-to-autosome diversity when compared to non-African populations. The arrows pointing to two orange dots indicate the sub-Saharan pygmy populations and the authors draw attention to how they have lower X-to-autosome diversity ratios than the other sub-Saharan African populations. They claim that this difference seen in pygmy populations could be due to demographic difference like male-driven admixture and not as much natural selection pressures.
Figure 1C: This is a heat-map of the the populations used in the SGDP and their estimated Neanderthal ancestry. The more yellow populations have the highest Neandethal ancestry values on a scale of 0-0.5% and those are seen mostly in East Asia, middle Neanderthal estimates (blue) in Europe,Eurasia, and the Americas and the lowest Neanderthal estimates (black) are seen in Africa. There is no Neanderthal ancestry seen in sub-Saharan population, and it begins to appear in the Northern African populations that were studied. The Australo-Melanesians appear to have fairly high estimates of Neanderthal ancestry.
Figure 1D: This is the same type of map with the exception that Denisovan ancestry is being estimated on the heat-map with a smaller scale of 0-0.5%. The smaller scale is used to bring attention to the difference in mainland Eurasia populations. Most of the global populations studied have little to no Denisovan ancestry detected through the SGDP estimates. However, there are high Denisovan ancestry levels in the Oceanian populations, and moderate level Denisovan ancestry in the Southeastern Asian populations.The data indicating Denisovan ancestry in South Asians has not been seen in previous studeis and could be because those studies did not include as many South Asian populations.
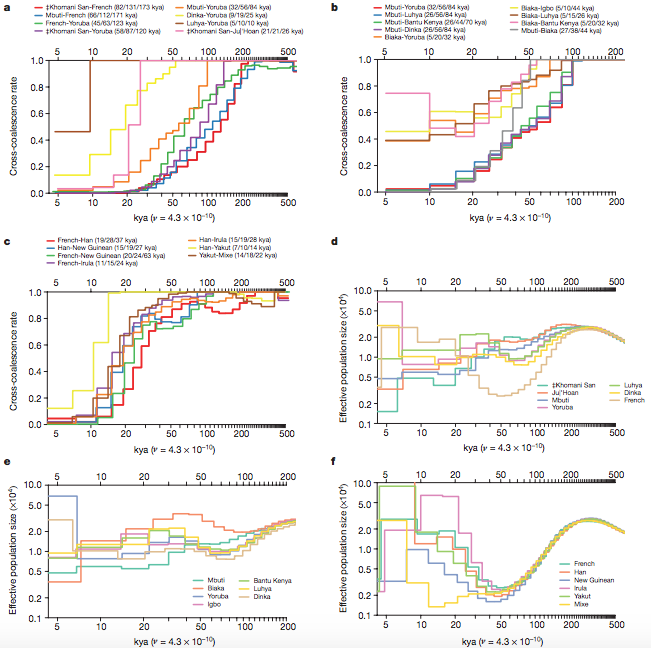
Figure 1A: By analyzing pairwise divergence per nucleotide in the data obtained from SGDP, the authors were able to construct a large neighbour-joining tree. These results agree with previous studies in that the deepest splits in human population evolution are between the African populations. The African populations are in the orange at the top of the tree and their branches split the farthest to the left, indicating that their populations diverged before any of the others that are included. Before forming this tree, the authors carried out studies on population relationships with ADMIXTURE and principle component analysis which both form inter-population relatedness reads using differences in genetic variability.
Figure 1B: This panel examines the heterozygosity of modern human populations from across the world by comparing the proportion of diallelic genotypes per base pair between populations.The data agrees with previous studies in that sub-Saharan African populations and pygymies (orange dots) have both the highest genetic diversity and ratio of X-to-autosome diversity when compared to non-African populations. The arrows pointing to two orange dots indicate the sub-Saharan pygmy populations and the authors draw attention to how they have lower X-to-autosome diversity ratios than the other sub-Saharan African populations. They claim that this difference seen in pygmy populations could be due to demographic difference like male-driven admixture and not as much natural selection pressures.
Figure 1C: This is a heat-map of the the populations used in the SGDP and their estimated Neanderthal ancestry. The more yellow populations have the highest Neandethal ancestry values on a scale of 0-0.5% and those are seen mostly in East Asia, middle Neanderthal estimates (blue) in Europe,Eurasia, and the Americas and the lowest Neanderthal estimates (black) are seen in Africa. There is no Neanderthal ancestry seen in sub-Saharan population, and it begins to appear in the Northern African populations that were studied. The Australo-Melanesians appear to have fairly high estimates of Neanderthal ancestry.
Figure 1D: This is the same type of map with the exception that Denisovan ancestry is being estimated on the heat-map with a smaller scale of 0-0.5%. The smaller scale is used to bring attention to the difference in mainland Eurasia populations. Most of the global populations studied have little to no Denisovan ancestry detected through the SGDP estimates. However, there are high Denisovan ancestry levels in the Oceanian populations, and moderate level Denisovan ancestry in the Southeastern Asian populations.The data indicating Denisovan ancestry in South Asians has not been seen in previous studeis and could be because those studies did not include as many South Asian populations.

Figure 2:
Cross-coalescence rates and effective population sized for selected
population pairs. (Malllick et al. 2016)
Figure 2A-C: Multiple sequentially Markovian coalescent (MSMC) was used to analyze time periods of human population divergences from Africa (a), Central African rainforest hunter-gatherers (b), and Ancient non-Africans (c). From panel A, the ancestry of present-day African hunter-gatherers indicates that the ancestral population to all present-day populations began developing around 200 thousand years ago. That is the time period when pretty much all of the cross-coalescence rates began to diverge in various ways. These panels also show estimates of time periods of when specific ancient populations may have diverged from each other, reflected by the separation of their rates of coalescence over time. This is how the authors produced rough estimates of when certain divergences occurred and which ones were most ancient and most recent. The most ancient non-African population was estimated to exist around 50 thousand years ago, much more recent than the ancient African population divergences.
Figure 2D-F: These panels are using the same populations as panels (a), (b), and (c), except pairwise sequential Markovian coalescent (PSMC) was used to estimate population size changes over time using one diploid genome per population. The authors do not discuss these panels in the text as much as the others, but the readers can notice how the populations sizes were fairly similar to each other in the more ancient times, and about 100 thousand to 50 thousand years ago the population sizes began to vary. These results were obtained through analyzing genetic variation in populations and their relations to each other, more specifically PSMC. Additionally, the human mutation rates are not definitive so estimates may not be extremely accurate.
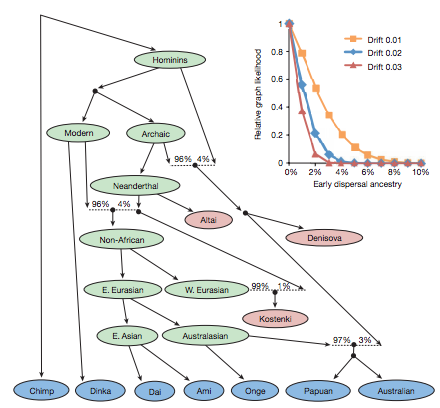
Figure 3: Present-day populations have negligible ancestry from an early dispersal of modern humans out of Africa.(Malllick et al. 2016)
The large model is an admixture graph showing the relationships between populations of various time periods and geographical locations. The present day populations are shown in blue, the ancient populations are in red, and the select inferred ancestral nodes are in green. Dotted lines are admixture events: when separate populations are breeding with each other. The phylogenetic tree was constructed using allele frequency correlations among the different population subgroups. The inset graph is showing the dispersal admixture to provide estimates of when the early lineage split off. The authors assigned the early lineage split to be just above the Non-African ancestral node. Each drift of 0.01 units represents 10,000 years associated with the genetic drift for that model. If the Oceanian and mainland East Asians populations branched off the main lineage forming non-Africans around 10-20 thousand years ago before the ancestors of European and East Asians diverged, there is a small percentage of ancestral contribution towards the Oceanian populations. These results oppose previous studies in suggesting that there is no impact of the earlier human dispersals on Oceanian populations, or any non-African population in general.
Reference List
Mallick, Li, Lipson, Mathieson, et al. 2016. The Simons Genome Diversity Project: 300 Genomes from 142 Diverse Populations. Nature 538:201-206.
Figure 2A-C: Multiple sequentially Markovian coalescent (MSMC) was used to analyze time periods of human population divergences from Africa (a), Central African rainforest hunter-gatherers (b), and Ancient non-Africans (c). From panel A, the ancestry of present-day African hunter-gatherers indicates that the ancestral population to all present-day populations began developing around 200 thousand years ago. That is the time period when pretty much all of the cross-coalescence rates began to diverge in various ways. These panels also show estimates of time periods of when specific ancient populations may have diverged from each other, reflected by the separation of their rates of coalescence over time. This is how the authors produced rough estimates of when certain divergences occurred and which ones were most ancient and most recent. The most ancient non-African population was estimated to exist around 50 thousand years ago, much more recent than the ancient African population divergences.
Figure 2D-F: These panels are using the same populations as panels (a), (b), and (c), except pairwise sequential Markovian coalescent (PSMC) was used to estimate population size changes over time using one diploid genome per population. The authors do not discuss these panels in the text as much as the others, but the readers can notice how the populations sizes were fairly similar to each other in the more ancient times, and about 100 thousand to 50 thousand years ago the population sizes began to vary. These results were obtained through analyzing genetic variation in populations and their relations to each other, more specifically PSMC. Additionally, the human mutation rates are not definitive so estimates may not be extremely accurate.

Figure 3: Present-day populations have negligible ancestry from an early dispersal of modern humans out of Africa.(Malllick et al. 2016)
The large model is an admixture graph showing the relationships between populations of various time periods and geographical locations. The present day populations are shown in blue, the ancient populations are in red, and the select inferred ancestral nodes are in green. Dotted lines are admixture events: when separate populations are breeding with each other. The phylogenetic tree was constructed using allele frequency correlations among the different population subgroups. The inset graph is showing the dispersal admixture to provide estimates of when the early lineage split off. The authors assigned the early lineage split to be just above the Non-African ancestral node. Each drift of 0.01 units represents 10,000 years associated with the genetic drift for that model. If the Oceanian and mainland East Asians populations branched off the main lineage forming non-Africans around 10-20 thousand years ago before the ancestors of European and East Asians diverged, there is a small percentage of ancestral contribution towards the Oceanian populations. These results oppose previous studies in suggesting that there is no impact of the earlier human dispersals on Oceanian populations, or any non-African population in general.
Reference List
Mallick, Li, Lipson, Mathieson, et al. 2016. The Simons Genome Diversity Project: 300 Genomes from 142 Diverse Populations. Nature 538:201-206.
https://www.nature.com/nature/journal/v538/n7624/full/nature18964.html