Overview:
b-lactamase is an enzyme whose activity allows for the in vivo quantitation of transcription when provided with a certain substrate. This enzyme is normally expressed through the ampicillin resistance gene, and its normal function is to cut the b-lactam ring of many antibiotics (e.g. penicillin or cephalosporins) (Sikorski and Peters 1998). The cutting of antibiotics has previously studied in depth in clinical settings due to the increasing rise of antibiotic-resistant strains of bacteria (van Belzen 1998). However, this cleaving also disrupts the normal activity of cellular fluorescence resonance energy transfer (FRET), which can be useful as a reporter system in cell cultures (Tsien and Miyawaki 1998). When a certain wavelength of light is shined onto the b-lactamase specific substrate inside the cells, the light reflected is of a different frequency, seen to the observer as different color.
There are three main advantages of using b-lactamase over other reporter methods:
![]() Active transcription is indicated
by a color change, not a change in brightness, and is thus easier
to detect activity (Zlokarnik et al.
1998).
Active transcription is indicated
by a color change, not a change in brightness, and is thus easier
to detect activity (Zlokarnik et al.
1998).
![]() The amount of b-lactamase
activity needed to detect transcription is 100 times less than
green fluorescent protein (Sikorski and
Peters 1998).
The amount of b-lactamase
activity needed to detect transcription is 100 times less than
green fluorescent protein (Sikorski and
Peters 1998).
![]() The ampicillin resistance gene
which encodes the enzyme is already in many developed plasmids
(Sutcliffe 1978), and cells just need
to be provided with the proper substrate.
The ampicillin resistance gene
which encodes the enzyme is already in many developed plasmids
(Sutcliffe 1978), and cells just need
to be provided with the proper substrate.
How b-Lactamase works:
The b-lactamase enzyme provides antibiotic resistance in Escherichia coli by rendering antibiotics harmless within the cell through the hydrolysis of penicillins to penicilloic acids (Sutcliffe 1978). b-lactamase will accumulate in the cytosol when its signal sequence is deleted (Kadonaga et al. 1984).
In the presence of a cephalosporin, b-lactamase will cleave the b-lactam ring, freeing an amino group, which in turn stimulates the elimination of a leaving group at the terminal end (Fig. 1) (Zlokarnik et al. 1998).
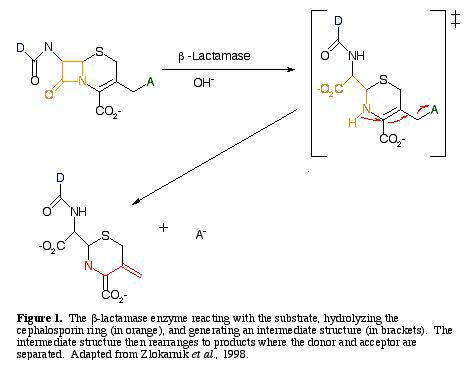
For a b-lactamase reaction to be detectable, the substrate must emit a short-wavelength donor from the long-wavelength acceptor when it is excited. The reaction disrupts intracellular FRET, and reestablishes fluorescence from the donor (Zlokarnik et al. 1998).
Zlokarnik et al. (1998) designed
a substrate, CCF2, to possess all of the characters needed in
a 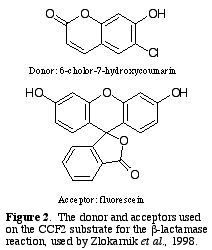 reaction with b-lactamase;
as well as an ester derivative of this substrate, CCF2/AM (AM=acetoxymethyl).
The donor fluorophore, D, is 6-chloro-7-hydroxycoumarin, and the
acceptor, A, is fluorescein (Fig. 2.).
reaction with b-lactamase;
as well as an ester derivative of this substrate, CCF2/AM (AM=acetoxymethyl).
The donor fluorophore, D, is 6-chloro-7-hydroxycoumarin, and the
acceptor, A, is fluorescein (Fig. 2.).
The substrate CCF2/AM is placed in contact with the cells. When CCF2/AM diffuses through the cell membrane, esterases within the cell hydrolyze the ester's functional abilities. The cleaved substrate, excited at 409 nm light, induces FRET within the cell, and emits 520 nm light (and green light is observed) (Fig 3A).
b-lactamase separates the acceptor from the donor, which disables FRET, and blue light is observed instead of green light (Fig. 3B).
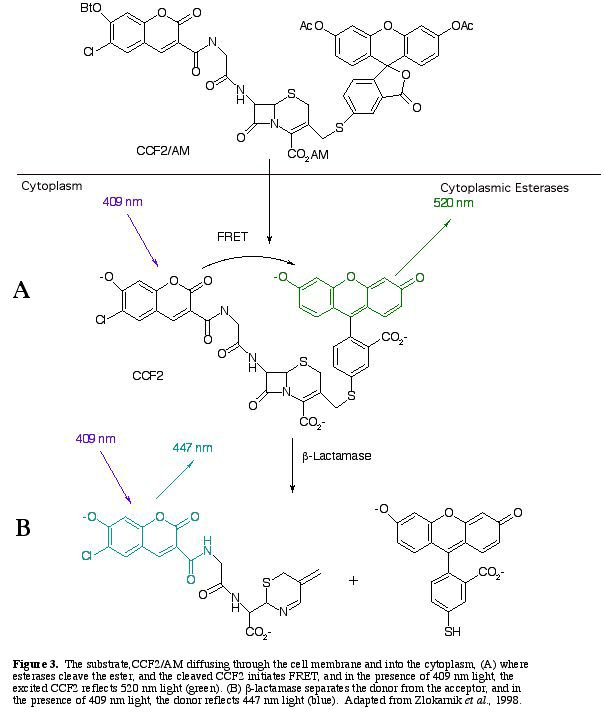
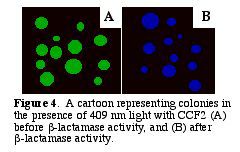
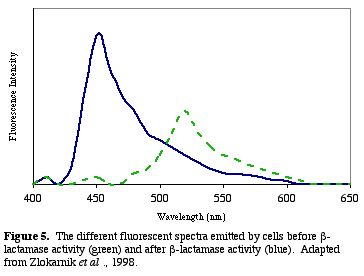
After b-lactamase cleaves CCF2, the donor glows blue with a fluorescent quantum yield of .74 (Zlokarnik et al. 1998). The blue within the cells can be distinguished visually (Fig. 4). A graph of the light intensities shows the different wavelengths of light emitted (Fig. 5.).
To spread b-lactamase throughout the entire cell, the protein can be tagged as a resident protein in the cytosol with an amino acid signal sequence of M-S or M-G (Kadonaga et al. 1984). As the enzyme is produced along with all other transcription in the cell, the level of blue observed in the cell is indicative of the amount of total transcription and translation occurring. With a single dosage of a reactive substrate, b-lactamase activity can be observed for up to 16 hours, as the enzyme has a determined half-life of 206 +12 min (Zlokarnik et al. 1998). Varying amounts of substrate provided can produce different intensities of blue, which can be optimized for specific application.
Several different strains of b-lactamases are available (e.g., OXA, TEM, and SHV) which use different substrates (van Belzen 1998). Some strains can be activated, or have increased activity with different reagents such as carbachol (Zlokarnik et al. 1998).
Different cell strains (e.g., Jurkat Cells, BHK cells, or HeLa cells) have different different characteristics of b-lactam breakdown (for a discussion of the different cell strains, see (Zlokarnik et al. 1998). A calibration curve for the specific cell line can indicate the amount of b-lactamase activity occurring within the cell.
b-lactamase technology takes advantage of a commonly used gene, and with the provided substrate, can give a clear indication as to the amount of genetic activity occurring. This reporter system will no doubt be of great use where sensitive quantitation of such activity is necessary.
![]() Kadonaga
JT, Gautier AE, Straus DR, Charles AD, Edge MD, Knowles JR. 1984.
Role of the beta-lactamase signal sequence in the secretion of
proteins by Escherichia coli. J. Biol. Chem. 259(4):
2149-2154.
Kadonaga
JT, Gautier AE, Straus DR, Charles AD, Edge MD, Knowles JR. 1984.
Role of the beta-lactamase signal sequence in the secretion of
proteins by Escherichia coli. J. Biol. Chem. 259(4):
2149-2154.
![]() Sikorski
R, Peters R. 1998. Lactamase Live! Science: 279 412.
Sikorski
R, Peters R. 1998. Lactamase Live! Science: 279 412.
![]() Sutcliffe
JG. 1978. Nucleotide sequence of the ampicillin resistance gene
of Escherichia coli plasmid pBR322. Proc. Natl. Acad.
Sci. USA 75(8): 3737-41.
Sutcliffe
JG. 1978. Nucleotide sequence of the ampicillin resistance gene
of Escherichia coli plasmid pBR322. Proc. Natl. Acad.
Sci. USA 75(8): 3737-41.
![]() Tsien
RY, Miyawaki, A. 1998. Seeing the machinery of live cells. Science
280: 1954-5.
Tsien
RY, Miyawaki, A. 1998. Seeing the machinery of live cells. Science
280: 1954-5.
![]() van
Belzen N. 1998 March. Beta-lactamase resources on the internet.
<http://www.md.huji.ac.il/biology/lactamase.html>.
Accessed 2000 Feb 20
van
Belzen N. 1998 March. Beta-lactamase resources on the internet.
<http://www.md.huji.ac.il/biology/lactamase.html>.
Accessed 2000 Feb 20
![]() Zlokarnik
G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng L, Whitney M,
Roemer K, Tsein RY. 1998. Quantitation of transcription and clonal
selection of single living cells with b-lactamase
as reporter. Science 279: 84-8.
Zlokarnik
G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng L, Whitney M,
Roemer K, Tsein RY. 1998. Quantitation of transcription and clonal
selection of single living cells with b-lactamase
as reporter. Science 279: 84-8.
Back to the Davidson College Molecular Biology Page
Back to Aaron Rice's Main Page
©2000 Department of Biology, Davidson College, Davidson, NC 28036
Questions, Comments? email: aarice@davidson.edu