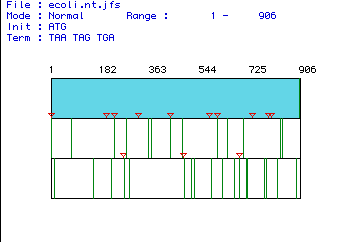
MacDNAsis Analysis of E. Coli Era (Ras Homolog) ORF

Figure 1. Depiction of complementary DNA sequence from E. coli Era gene. The largest ORF lies in the first reading frame and spans the entire region from nucleotide 1-906. Original nucleotide sequence: E. Coli sequence Predicted molecular weight of E. Coli Era: 33808 Daltons.
Predicted Characteristics of Era
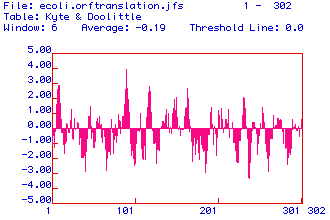
Figure 2. Hydrophibicity plot suggestive of four possible transmembrane
domains where plot values exceed 2.00. Therefore, Era is possibly
an integral membrane protein.
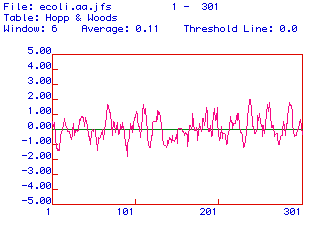
Figure 3. An important characteristic of epitopes is that they
are exposed in order to interact with antibodies when the molecule is in
its native conformation. Oligopeptides from regions where the plotted
values are continuously over 0.00 such as from amino acids 40-60 or 220-240
are predicted to be hydrophilic and consequently would be suitable for
antibody generation.
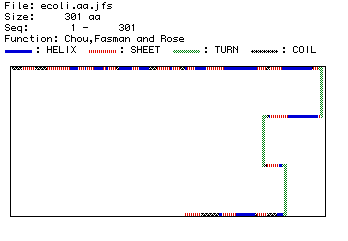
Figure 4. Predicted secondary structure of Era.
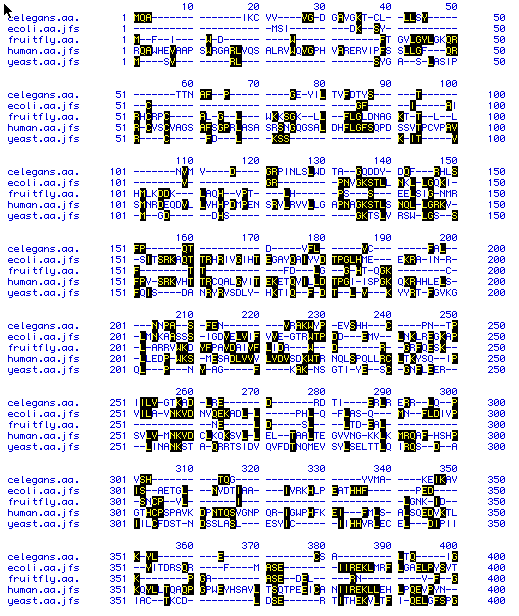
Figure 5. Gapped alignment by Higgins method. Conserved
amino acids are highlighted in black. Links to: C.
elegans, E. coli, D.
Melanogaster, H. sapiens, S.
cerevisiae amino acid sequences.
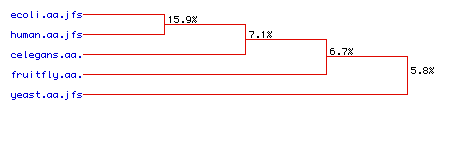
Figure 6. Phylogenic tree for Ras homologs. Representation of genetic distance between species. Links to: C. elegans, E. coli, D. Melanogaster, H. sapiens, S. cerevisiae amino acid sequences.
[Davidson Molecular Biology web page]
Email any questions, comments or suggestions to: fisturgill@davidson.edu