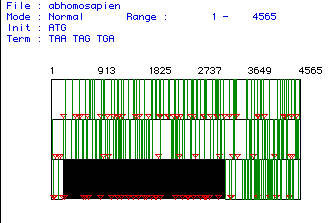
Using the version 3.5 MacDNAsis program, various analyses of the cDNA for phospholipase C were performed.
I: Open Reading Frame (ORF)
The largest open reading frame of the cDNA for phospholipase
C was found. This segment of the DNA began at
nucleotide number 204 and ended at nucleotide number 3197. The image below
shows the entire segment of cDNA. The largest reading
frame, indicated by the black box, was the segment of DNA used for the
remainder of the DNA analysis.

Figure #1: Determination of the Largest Open Reading Frame (ORF). This image depicts the largest open reading frame for the phospholipase C cDNA isolated from humans. The nucleotide numbers are listed above the reading frames. Red triangles indicate start codons (AUG) and vertical lines represent stop codons. The largest open reading frame was found starting at nucleotide 204 and ending at nucleotide 3197.
II. Translation and Determination of the
Predicted
Molecular Weight
The ORF obtained above was translated (DNA to protein)
and the predicted
molecular weight was then determined to be 112,
949.62 daltons.
III. Kyte and Doolittle Hydropathy Plot
Using the algorithm constructed by Kyte and Doolittle,
a hydropathy plot was used
to predict if phospholipase C is an integral membrane
protein. The plot is shown below:
Figure 2: Kyte and Doolittle Hydropathy Plot. This plot was made using the algorithm constructed by Kyte and Doolittle. A reading of greater than or equal to +1.8 indicates an area of the protein that has enough hydrophobicity to be an integral membrane protein. No peak on this plot has a hydrophobicity greater than or equal to +1.8, thus indicating that no transmembrane domains are present.
Hydrophobic areas of the protein are indicated by positive values and hydrophilic areas are indicated by negative values on this plot. A transmembrane domain is only present if a segment of the protein has a hydrophobicity reading greater than or equal to +1.8. As shown in this plot, no segments of phospholipase C have a hydrophobicity greater than or equal to +1.8. Therefore, this plot predicts that phospholipase C is not an integral membrane protein.
IV. Hopp and Wood Antigenicity Plot
Below is an antigenicity plot constructed using the algorithm
developed by Hopp and Wood:
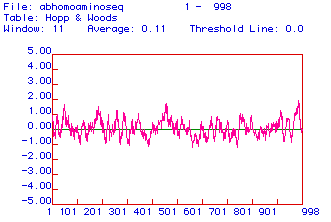
Antigenic plots are used to predict the location of charged, hydrophilic areas of the protein. Charged areas of the protein direct themselves away from the hydrophobic phospholipid bilayer of the plasma membrane and are most likely to be found in the cytoplasm. These hydrophilic areas of the protein can be detected and bound by monoclonal antibodies. In the antigenic plot above, hydrophilic areas of phospholipase C predicted to be used for the production of monoclonal antibodies are located around amino acid 50, 450, and the last 20 amino acids.
V. Prediction of the Secondary Structure of Human
Phospholipase C
The translated ORF of human phospholipase C cDNA was also
analyzed through
contruction of a predicted secondary structure map:
This predicted secondary structure was compared with the crystallized structure of phospholipase C. To view the Rasmol image of phospholipase C, click here.
The predicted secondary structure from the MacDNAsis does not agree with the actual three dimensional structure seen in the RasMol image.
VI. Multiple Sequence Alignment
In this section of the MacDNAsis analysis, the phospholipase C protein sequences from Bos taurus (bovine), Drosophila melanogaster (fly), Homo sapien (human), Rattus norvegicus (rat), and Saccharomyces cerevisae (yeast). A short sequence of the comparison of the amino acid sequences is shown below.
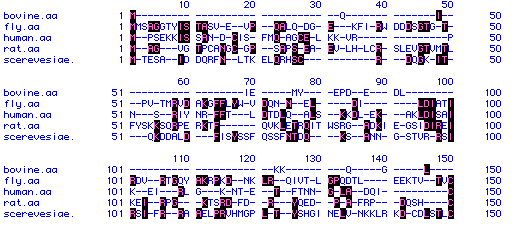
Figure 5: Multiple Sequence Alignment. A multiple sequence alignment was constructed on the amino acid sequences of phospholipase C from the five organisms listed above (Bos taurus, Drosophila melanogaster, Homo sapien, Rattus norvegicus, and Saccharomyces cerevisae). Only part of the alignment is shown above. Amino acid residues with black boxes indicate those residues that appear in more than one organism. Dashes (-) indicate spaces that were introduced to better align the five different phospholipase C sequences.
VII. Phylogenetic Tree
A phylogenetic analysis was performed to quantitatively compare the multiple sequence alignments of phospholipase C. The purpose of this analysis was to determine the degree of amino acid conservation. The percentages seen in the phylogenetic tree below display the degree of similarity between the amino acid sequences of the five species.
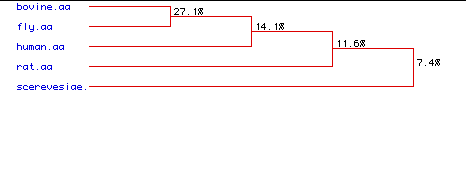
Figure 6: Phylogenetic Tree. A phylogenetic analysis was performed to find the degree of amino acid conservation between the five types of phospholipase C. Percentages indicate the degree of similarity between the amino acid sequences of the five species.
In the phylogenetic tree above, it is interesting to note that a greater degree of similarity is seen between the bull and the fly than between the bull and the human. One would predict that there would be a greater degree of amino acid conservation between two mammals than between a bull and a fly. Overall, the degree of similarity between the five species is low.