This web page was produced
as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Protein
RAS1
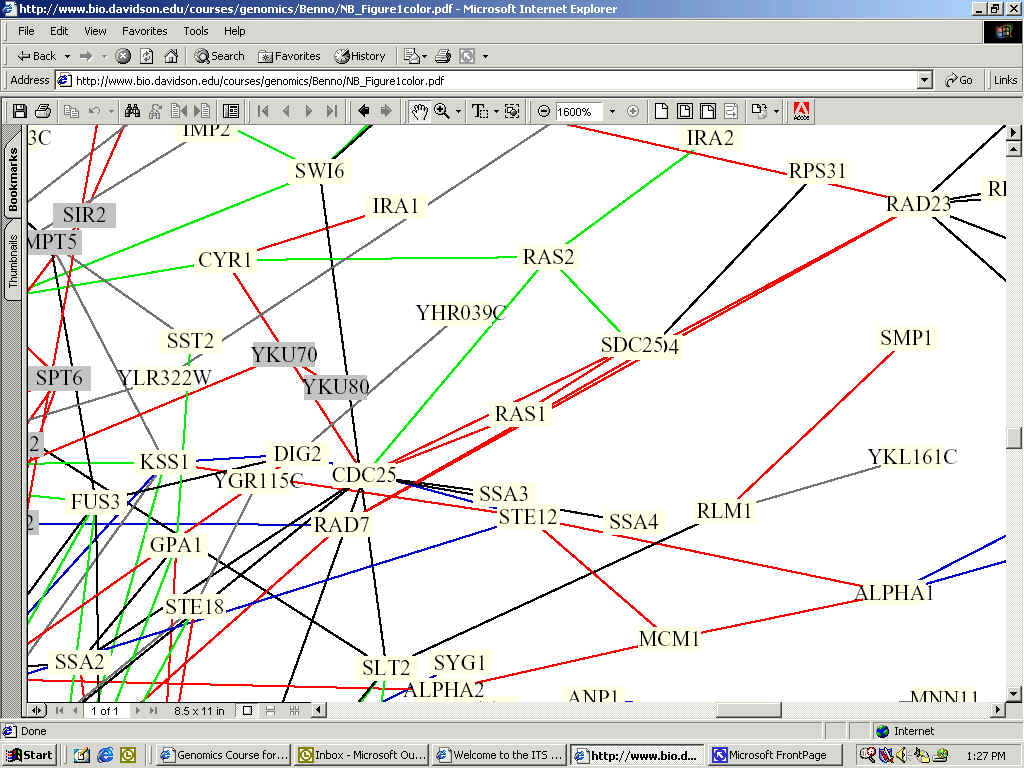
In identifying the function of my favorite yeast gene's encoded protien, there are many databases and experiments to draw upon in order to gain further insights into its function and interaction with other proteins in yeast cells. Below is a portion of a synthesized interaction map for yeast compiled by Stan Fields, and Peter Uetz, aided by Benno Schwikowski, a computer science expert at the Institute for Systems Biology. This one integrated circuit shows the relationship between 2,039 different proteins (a small portion of which is shown). RAS1 is almost directly center.

Full Image: http://bio.davidson.edu/courses/genomics/Benno/NB_Figure1color.pdf
The
color key for the lines is: red = cellular
role and subcellular localization of interacting proteins are identical;
The figure shows that RAS1 interacts with multiple proteins, specifically CDC25, SDC25, RAD7, and another protein whose name is blocked. Because all of the lines connected to RAS1 are red, the cellular role and subcellular localization of interacting proteins are identical.
Searching FunctionJunction resulted in the following descriptions of the related proteins:
CDC25: Blocks cell division cycle at 36 degree C. Involved in RAS protein signal transduction, cell cycle control, and in the start point of mitotic cell cycle. Located in cytoplasm and plasma membrane.
SDC25: RAS protein signal transduction. Otherwise, biological process and cellular component unknown.
RAD7: Repairosome, involved in nucleotide-excsision repair.
CDC25's role of blocking cell division at 36 degrees C is complementary to the function of RAS1 maintaining homeostasis in response to stress. When RAS1 is activated because of unfavorable conditions, it begins controlling cell growth by interacting with CDC25. Another interesting factor of CDC25 is its association with the mitotic cell cycle. This indicates that RAS1 is able to signal to CDC25 whether or not it should allow mitosis to begin. According to work by Mintzer and Field, Cdc25 is the guanine nucleotide exchange protein that activates Ras. Ras, in turn, activates adenylyl cyclase.
SDC25, as a RAS protein signal transduction protein, seems to serve to communicate and coordinate activities between RAS1 and RAS2, both of which are involved in the cell aging process. Again, the fact that it interacts with RAS1 makes sense.
Finally, the relation to RAD7 further supports the role of RAS1 in responding to stress. If RAS1 recognizes that a stressful situation has occured and possibly altered the genetic code, it signals RAD7 to repair the damage, thus helping to maintain and determine life span.
In the following integrated circuit, Fields et al. show the connections between proteins associated with aging and other proteins. Again, RAS1 is almost directly centered and now colored Indian Red:
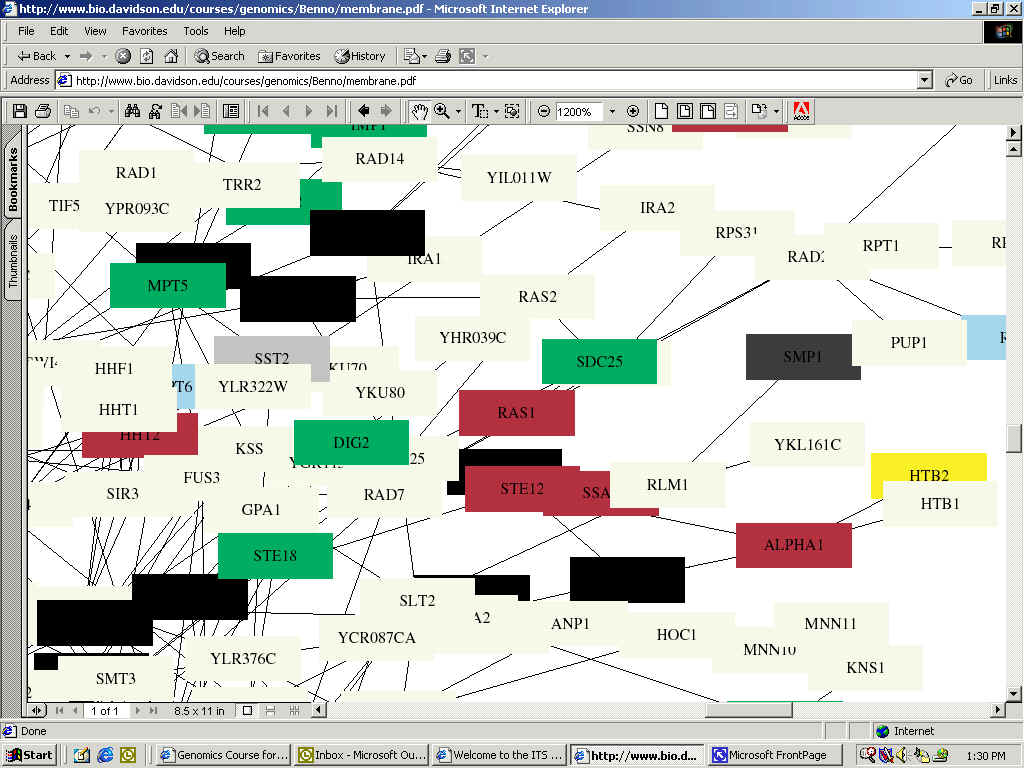
(Indian Red Color indicates function of small-molecule transport;.Green: Cell-cycle Control;Black: RNA-Turnover; Gray: Membrane Fusion; Light Blue: Protein Folding)
Full Image: http://bio.davidson.edu/courses/genomics/Benno/aging.pdf
In the above figure, we find the same relationships as in the first interaction map, as well as other possibilities which are impossible to decipher due to being blocked. Here, we are also told that SDC25, described above to be involved in signaling between RAS1 and RAS2, plays a role in cell-cycle control, as indicated by its green color. RAS1 needs to be able to alter and influence cell cycle in order to fully establish a homeostatic balance as it is thought to do (for background details of RAS1's known functions, please refer to my home page).
In order to further investigate the proteins that interact with RAS1, the Database of Interacting Proteins (DIP) was consulted. Although the site provides excellent java-enhanced figures of protein interactions, the figure below, a static figure, shows the protein interactions with RAS1.

Here, RAS1 is the central node (red). Coming off of this node going clockwise from the top are AKR1, CDC25, S59828, MEK1, JQ1027, and BYR2_Schpo. Although we were already aware of the interaction with CDC25, the rest are new protein interactions to be explored. It is interesting to note the differences between the results from this website and the results from the pdf files from experimentation above. There exists no answer as to which is a better prediction, but it is more likely that because of the complex nature of RAS1, it is involved in many different pathways. Below are the protein properties for the newly discovered interactions according to FunctionJunction:
MEK1: Involved in meiosis and protein amino acid phosphorylation and is located in the nucleus. Required for full double strand breaks, normal length synaptonemal complexes, meiotic recombination, and spore viability.
AKR1: Involved in endocytosis and signal transduction of mating signal with unknown location in the cell. Negative regulator of pheromone response pathway; required for endocytosis of pheromone receptors and involved in cell shape control.
S59828, JQ1027, and BYR2_Schpo were not in FunctionJunction's database.
RAS1's relationship with MEK1 is somewhat surprising. Although RAS1 plays a role in cell proliferation, and as a cell regulator in general would need to be involved in protein amino acid phosphorylation, its hypothesized location in the plasma membrane would lead me to believe that it would not be able to directly interact with MEK1 which is in the nucleus. Interestingly, in a study by Van Aelst et al., it was found that RAS1 and MEK1 only interact when RAF, another oncoprotein, was not present. AKR1 is very similar to RAS1 as they are both involved in signal transduction. Again, if RAS1 is going to regulate the cell, it also needs to be able to regulate the pheromone secretion, in turn controlling proliferation.
Consulting Curagen Corporation's PathCalling, which finds protein interactions, CDC25 and SDC25 once again appeared as being related to RAS1.
Along with associated proteins, the structure of the RAS1 can also serve to give insight into its function, as similar structure can be more revealing than similar nucleotide sequences in some cases. When visiting the PSIPRED database, which also can be used to do GenThreader and MEMSAT searches, a large amount of information was discovered. First, it is helpful to look at an ortholog of RAS1, the human Ras gene:
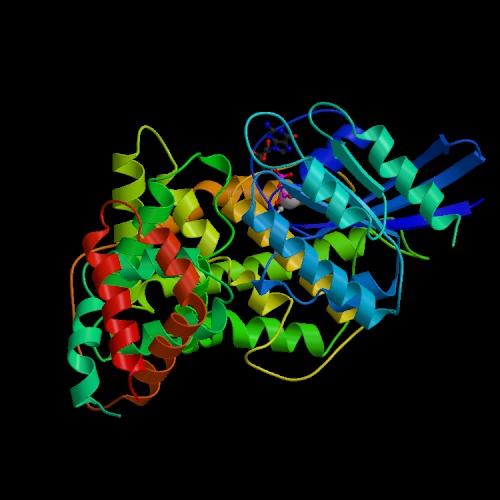
This image was taken from Structure Explorer. Permission Pending.
In analysing RAS1 in particular, the first step was to determine the predicted structure of the protein using PSIPRED. This program gives a graphical output which can be accessed in PDF format here. This showed the suspected location of strands, helices, and coils in the RAS1 protein. This same amino acid sequence was then put through the MEMSTAT program to locate transmembrane helices, of which RAS1 had none. Finally, GenThreader was used in order to determine similar structures to RAS1. Using this program, a number of previously crystalized structures were returned as being similar. Although these are all from different organisms, they provide insight and confirmation into the role of RAS1. The most similar PDB ID, is 1ctq, a signaling protein, which has the following structure:
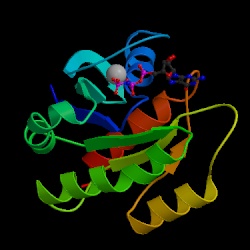
This image was taken from Structure Explorer. Permission Pending.
This further reinforces the idea that RAS1 is a signaling protein, having a .96 probability of being very similar to the above structure. The other ID's resulted in structures representing GTP-binding proteins, transport proteins, and other proteins with similar functions to RAS1. One unusual similarity was what with PDB ID: 1d2e which is an RNA binding protein. However, this is the last of the ten similar structures reported by the program. Again, the hypothesized function of RAS1 is supported by the structure of similar molecules.
Because RAS1 was the first oncogene discover, a great deal is known about both its structure and function. Therefore, it is not surprising that my research did not come up with new or unusual information. However, the methods I followed, and the sites explored provide an excellent starting place in trying to discern the function of a protein based on related proteins and structure.
YMR086W
Unlike RAS1, there was not an abundance of information about YMR086W available. In fact, none of the sources used for RAS1 reported any proteins that YMR086W interact with, nor was any significant structural relationship found. In fact, the most similar structure found using Structure Explorer, with a similarity probability of .13, was simply intertwined ribbons. Using GenThreader, PSIPRED, and MEMSTAT, some information was discovered, but it did not further aid in understanding the function of YMR086W. Interestingly however, the MEMSTAT result, which can be seen here, showed that YMR086W does have a transmembrane domain, something which the Kyte-Doolittle analysis performed earlier had dismissed. The pdf for the GenThreader structure prediction can be seen here, although it does not reveal much about the structure of YMR086W, with only two helices and one strand. One new piece of information, according to the Gerstein Lab at Yale, is that YMR086W is thought to be located in the nucleus.
So, since not much is known about the function of YMR086W, it is necessary to design experiments to try and identify its function. These experiments should take into account previously discovered (hypothesized) relationships. From the microarray analysis performed earlier, it was proposed that YMR086W may be involved with protein production and cell cycle activity.
To test these hypotheses there are two main methodologies: looking at the protein's structure, and looking at the protein's relationship with other proteins. Unfortunately, without crystalizing the protein, we are unable to attain its structure. Over time, however, it is possible that other similar structures will be crystalized, rendering the information known about YMR086W useful. Otherwise, with the hypothesis of involvement with protein production and cell activity, we would try and test whether or not YMR086W interacts with any other proteins that are known to be involved in protein production and cell cycle activity. Below, I have designed two experiments to try and attain this information.
EXPERIMENT 1:
The first experiment will utilize the yeast two-hybrid method (Y2H) developed by Stan Fields. This method will use YMR086W as a bait in order to find proteins that interact with the bait protein, known as the prey. This method works as follows: A transcription factor is split into two parts, the DNA Binding Domain (DBD) and the Activation domain (AD) which tells the RNA polymerase to start transcription. Attached to the DBD is YMR086W (or any bait of interest), which cannot initiate transcription on its own. Fused to the AD side is the prey protein, which also cannot initiate transcription alone. When the two fused proteins are produced by the same cell, transcription of the reporter gene (any necessary gene that will determine life or death of cell) will occur if the bait and prey interact (Campbell 463). In this way YMR086W can be tested against every protein in the proteome, and interaction is shown by growth on a medium lacking the product encoded by the reporter gene. For a control, each prey must be grown in the absence of bait.
Using YMR086W as the bait, hundreds of prey proteins could be tested, beginning with those that are involved in protein production and cell cycle activity. However, it is still necessary to test against many different proteins as the exact function of YMR086W is unknown, and could possibly be very complex. This method would work well for YMR086W because so little is known about it, but there is also a starting point with the protein production and cell cycle control proteins. It is important to note that just because YMR086W binds to a protein does not necessarily mean that they interact. It is possible that they are never present together during cell cycles, or that they never come into contact with eachother in reality. Although this method is very laborious, it will provide excellent insight into the functioning of YMR086W, and could serve to accurately predict its function within yeast. However, if my hypothesis is correct that YMR086W is involved in protein biosynthesis, and this occurs at an early stage (as would be signified by its proposed location in the nucleus), then there could be a number of false positive interactions.
EXPERIMENT 2:
Another way to find the function of YMR086W would be to use a more complex method of attaining proteomic interaction. Gavin MacBeth and Stuart Schreiber from Harvard University created protein arrays that "covalently attach to a glass microscope slide one protein of a known pair to create a type of bait similar to the two-hybrid system (Campbell 497)". In this experiment in particular, YMR086W would be labled with a color, say red, and tested against a microarray with up to 10,800 other proteins spotted out to see where YMR086W bound. Like in the above experiment, those proteins that interact with YMR086W will 'stick' to it, and hence be colored red. By identifying which genes react with YMR086W, you can predict its function more accurately.
This high through-put method is probably a better way of testing YMR086W because of the uncertainty surrounding its functioning. This would allow for a quick look at which proteins it interacts with, and can use a wide array of protein types. MacBeth and Schreiber have tested its functionality quite extensively, and shown it to be quite accurate in proper binding. However, some of the same reservations about presence of binding proteins concurrently with YMR086W are still questionable. Following the results of the above experiment, if a number of false positives are thought to have occurred, then this experiment should be set up specifically to match up with mRNA splicing proteins and transcription factors. This would allow pin-pointing of YMR086W's functioning.
CONCLUSION
With these two experiments, and the resulting information that will be deciphered once protein relationships are known, the function of YMR086W will be better understood. I predict that YMR086W is located in the nucleus and builds proteins that in turn control cell cycle. Because it is in the nucleus, YMR086W is most likely either involved in mRNA splicing or is a transcription factor.
Campbell, A. Malcolm. "Chapter Six: Proteomics." Genomics, Proteomics, and Bioinformatics. 2001.
"Database of Interacting Proteins." UCLA. 1999. <http://dip.doe-mbi.ucla.edu/dip/represent_JAVA.cgi?PKEY=1041> 11/04/01.
"Function Junction." Saccaromyces Genome Database. 2001 <http://genome-www.stanford.edu/cgi-bin/SGD/functionJunction> 11/04/01.
Gerstein Lab. Yale University. 2001. <http://bioinfo.mbb.yale.edu/genome/yeast/search.cgi?orf=YMR086W> 11/07/01.
Mintzer KA, Field J. "The SH3 domain of the S. cerevisiae Cdc25p binds adenylyl cyclase and facilitates Ras regulation of cAMP signalling." Department of Pharmacology, University of Pennsylvania School of Medicine. <http://www.ncbi.nlm.nih.gov/htbin-post/Entrez/query?uid=0010048790&form=6&db=m&Dopt=b>
Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. "Complex formation between RAS and RAF and other protein kinases." Cold Spring Harbor Laboratory, NY. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8327501&dopt=Abstract>
"PathCalling." Curagen Corporation. Nov 2001. <http://portal.curagen.com/extpc/com.curagen.portal.servlet.PortalYeastGene?geneIdIn=4492&keywordIn=YMR086w> 11/04/01.
Protein Data Bank. 2001 <http://www.rcsb.org/pdb/index.html> 11/04/01.
Proteome Database. "YPD." 2001<http://www.proteome.com/databases/YPD/YPDsearch-quick.html> 11/04/01.
"Quick Search." Saccaromyces Genome Database. 2001. http://genome-www4.stanford.edu/cgi-bin/SGD/search/quickSearch?query=byr2_schpo 11/04/01.
Structure Explorer. "Ras-Rasgap Complex." Protein Data Bank. 2001. <http://www.rcsb.org/pdb/cgi/explore.cgi?job=graphics;pdbId=1WQ1;page=0;pid=293041005074537&opt=show&size=500> 11/04/01.
Davidson College Biology Department
© Copyright 2001 Department of Biology, Davidson College, Davidson, NC 28036