This web page was produced as an assignment for an
undergraduate course at Davidson College.
My Favorite Yeast Genes
These genes can be found in brewer's yeast, Saccharomyces cerevisiae.
For more background information on S.cerevisiae visit the
SGD "What are yeast?" page or the
Wyeast Laboratory educational page. For more information on the
genetics of this yeast species visit the Saccharomyces
Genome Database.
UBI4
UBI4 (ACC#:
Z73144), also known as SCD2 or the polyubiquitin gene, is found
on chromosome 12 of S.cerevisiae. The locus consists of 6209
base pairs (A: 1806, T: 2086, C: 1218, G: 1099) which code for 1360
amino acids (ACC#:
CAA97490.1). NCBI, 2001
Ozhaynak et al. reported that UBI4 "encodes a polyubiquitin precursor protein
containing five ubiquitin repeats in a head-to-tail, spacerless arrangement."
Ozkaynak,
1987; Finley, 1987 However, NCBI reports that the UBI4
encoded protein has no conserved domains. NCBI,
2001; <http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi>
UBI4 is related to UBI1, UBI2, and UBI3 which "encode hybrid proteins in
which ubiquitin is fused to unrelated ('tail') amino acid sequences." Ozkaynak,
1987 UBI4 encodes for ubiquitin, a protein found in the
cytoplasm of S.cerevisiae, similar to the proteins ubiquitin and polyubiquitin
found in rats and humans. SGD, 2001; <http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=YLL039c>,
<http://genome-www.stanford.edu/cgi-bin/SGD/Sacch3D/getblast?name=UBI4&db=mammal>
UBI4's protein is involved in the processes of "deubiquitylation, monoubiquitylation,
polyubiquitylation, sporulation (sensu Saccharomyces), and stress
response." Its biochemical activity is "protein degradation tagging"
but the gene is only expressed under conditions of stress.
SGD, 2001; <http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=YLL039c>
Numerous studies link UBI4 expression to the S.cerevisiae stress response
system which can be activated by heat shock elements (HSEs) and/or stress
response elements (STREs). Arnason, 1994;
Barbet, 1996; Cheng, 1994; Finley, 1987; Fraser, 1991; Hazell, 1995; Lee,
1996; Ozkaynak, 1987; Simon, 1999; Tanaka, 1988; Timblin, 1997; Treger,
1988; Watt, 1997 According to Ozhaynak et al., "conjugation
of ubiquitin to intracellular proteins mediate their selective degradation
in vivo." Ozkaynak, 1987
In December 1988, Tanaka et al. proposed "that the UBI4 gene is one of
the genes which are part of the cAMP-effector pathway and required for
G-0G-1 arrest in Saccharomyces cerevisiae." Tanaka,
1989
There is a mutant allele for UBI4, ubi4, caused by systematic deletion
of the UBI4 gene. Winzeler, 1999
Finley et al. describe the mutant phenotypes: "ubi4 deletion mutants are
viable as vegetative cells, grow at wild-type rates, and contain wild-type
levels of free ubiquitin under exponential growth conditions. However,
although ubi4/UBI4 diploids can form four initially viable spores, the
two ubi spores within the ascus lose viability extremely rapidly, apparently
a novel phenotype in yeast. Furthermore, ubi4/ubi4 diploids are sporulation-defective.
ubi4 mutants are also hypersensitive to high temperatures, starvation,
and amino acid analogs. These three conditions... are all known to
induce stress proteins." Finley, 1987
SGD's description of ubi4 mutant phenotypes matches that published by Finley
et al. SGD, 2001; <http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=YLL039c>
SET4
SET4 (ACC#:
249380.1) is found on chromosome 10 of S.cerevisiae. The
locus consists of 3999 base pairs (A: 1307, T: 1216, C: 735, G: 741) which
code for 560 amino acids (ACC#:
CAA89400). SGD, 2001; <http://genome-www2.stanford.edu/cgi-bin/SGD/getSeq?map=nmap&seq=YJL105W&flankl=0&flankr=0&rev=>,
<http://genome-www2.stanford.edu/cgi-bin/SGD/getSeq?map=pmap&seq=YJL105W&flankl=0&flankr=0&rev=>
GeneMark
reports that SET4 has 1683 base pairs, with a CG value of 37.85%. GeneMark,
2001; <http://opal.biology.gatech.edu/GeneMark/> SET4
shares conserved domains labeled "PHD" and "SET" with other genes; the
e-values of these matches are low, 1e-6 and 4e-25
respectively, so these matches are probably not due to chance. PHD
tends to form zinc fingers. NCBI describes SET domains: "SET domains
appear to be protein-protein interaction domains. It has been demonstrated
that SET domains mediate interactions with a family of proteins that display
similarity with dual-specificity phosphatases (dsPTPases). A subset of
SET domains have been called PR domains. These domains are divergent in
sequence from other SET domains, but also appear to mediate protein-protein
interaction... SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain;
Putative methyl transferase, based on outlier plant homologues." NCBI,
2001; <http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi>
The program, PREDATOR,
can predict the secondary structure of a protein. The figures below
show the PREDATOR graphical output for SET4.
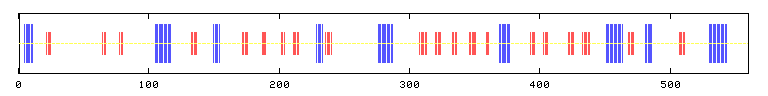
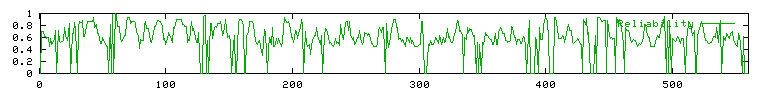
Figures 1A and 1B: X-axis refers to amino acid count.
Top figure shows secondary structure predicted states, where blue marks
represent alpha helices, red marks represent extended strands and yellow
marks represent random coil regions. Bottom figure shows predicted
reliability of state predictions.
According to PREDATOR, SET4's 560 amino acids can be devided into nine
regions of alpha helices (15%) and twenty regions of extended strand (16.07%)
interspersed with regions of random coil (68.93%). The reliability
of these predictions ranges. When the graph reaches or exceeds the
value 0.8 on the Y-axis of the reliability graph the secondary structure
state predictions in the upper graph are probable. The reliability
of the predicted secondary structure of SET4 is fairly high. These
results imply that SET4 may code for an integral membrane protein, like
a G protein-coupled receptor, since it contains many regions of alpha helix.
A hydropothy
plot, also known as Kyte-Doolittle analysis, predicts whether or not
a protein is an integral membrane protein. If peaks occur at or above
the value 2 on the graph then there is potential for a transmembrane domain.
The following figure is a hydropothy plot for SET4.
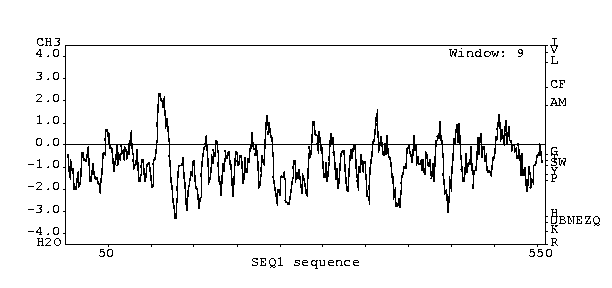
Figure 2: Courtesy of J. Kyte and R. F. Doolittle (1982)
J. Mol. Biol. 157:105-132. X-axis refers to amino acid count.
Graph represents intermembrane domain potential.
According to the hydropothy plot, there is only one section of the protein
encoded by SET4 which has high potential to be a transmembrane domain.
This domain occurs approximately at amino acid 100 of the sequence.
It should be noted that the PREDATOR secondary structure predictions show
a region of alpha helix that between approximately amino acid 100 and amino
acid 115 of the sequence.
Sources Consulted:
-
Arnason, Terra and Michael J. Ellison. (1994) "Stress resistance
in Saccharomyces cerevisiae is strongly correlated with assembly of a novel
type of multiubiquiton chain." Molecular and Cellular Biology
14(12): 7876-7883.
-
Barbet, Nik C., Ulrich Schneider, Stephen B. Helliwell, Ian Stansfield,
Michael F. Tuite, and Michael N. Hall. (1996) "TOR controls translation
initiation and early G1 progression in yeast." Molecular Biology
of the Cell 7(1): 25-42.
-
Cheng, L. R.Watt, and P.W.Piper. (1994) "Polyubiquitin gene expression
contributes to oxidative stress resistance in respiratory yeast." Molecular
and General Genetics 243(3): 358-362.
-
Finley, D., E.Ezkaynak, and A.Varshavsky. (1987) "The yeast polyubiquitin
gene is essential for resistance to high temperatures, starvation, and
other stresses." Cell 48(6): 1035-1046.
-
Fraser, J., H.A.Luu, J.Neculcea, D.Y.Thomas, and R.K.Storms. (1991) "Ubiquitin
gene expression: Response to environmental changes." Current Genetics
20(1-2): 17-24.
-
"GeneMark." GeneMark, 2001. <http://opal.biology.gatech.edu/GeneMark/>
-
"Genomic Sequence Total Analysis and Lookup Tool." GESTALT, 2001.
<http://sun01.systemsbiology.net/~gglusman/GESTALT/>
-
Hazell, Brian W., Helena Nevalainen, and Paul V. Attfield. (1995) "Evidence
that the Saccharomyces cerevisiae CIF1 (GGSI/TPSI) gene modulates heat
shock response positively." FEBS Letters 377(3): 457-460.
-
Kyte, J. and R.F.Doolittle (1982) J. Mol. Biol. 157:105-132.
-
Lee, Jinhwa, Annette Romeo, and Daniel J. Kosman. (1996) "Transcriptional
remodeling and G-1 arrest in dioxygen stress in Saccharomyces cerevisiae."
Journal
of Biological Chemistry 271(40): 24885-24893.
-
"National Center for Biotechnology Information." NCBI, 2001.
<http://www.ncbi.nlm.nih.gov/>
-
Ozkaynak, E., D.Finley, M.J.Solomon and A.Varshavsky. (1987) "The
yeast ubiquitin genes: A family of natural gene fusions." UMBO
Journal 6(5):1429-1440.
-
"Saccharomyces Genome Database." SGD, 2001. <http://genome-www.stanford.edu/Saccharomyces/>
-
Simon, John R., Janet M. Treger, and Kevin McEntee. (Feb. 1999) "Multiple
independent regulatory pathways control UBI4 expression after heat shock
in Sacharomyces cerevisiae." Molecular Microbiology 31(3):
823-832.
-
Takana, K., K.Matsumoto and E.A.Toh. "Dual regulation of the expression
of the polyubiquitin gene by cyclic AMP and heat shock in yeast." UMBO
Journal 7(2):495-502, 1988.
-
Timblin, Barbara K., and Lawrence W. Bergman. (Dec.1997) "Elevated
expression of stress response genes resulting from deletion of the PHO85
gene." Microbiology 26(5): 981-990.
-
Treger, J.M., K.A. Heichman, and K.McEntee. (1988) "Expression of the yeast
UBI4 gene increases in response to DNA-damaging agents and in meiosis."
Molecular and Cellular Biology 8(3): 1132-1136.
-
Watt, R. and P.W.Piper. (1997) "UBI4, the polyubiquitin gene of Saccharomyces
cerevisiae, as a heat shock gene that is also subject to catabolite depression
control." Molecular and General Genetics 253(4): 439-447.
-
Winzeler EA, et al. (1999) "Functional characterization of the S. cerevisiae
genome by gene deletion and parallel analysis". Science 285(5429):901-906.
updated 10/7/01


