Expression of Saccharomyces’ ORF HO
The ORF HO is an endonuclease that
mediates the switch of genes at the MAT locus with those at the HMR locus
during Saccharomyces’ mating type switch (Kuplun
2000). A search for the ORF HO on the
Saccharomyces Genome Database's Expression Connection outlines the expression
data for HO in microarray experiments involving HO's expression depending on alpha-factor concentration, in response to
alpha-factor, during the diauxic shift, during sporulation, in response to
histone depletion, and during the cell cycle. Data gathered in microarray
experiments should reflect the hypothesis that the ORF is involved in mating
type switching and thus repressed when exposed to pheromones and during
sporulation.
Each experiment
uses this scale to represent induction or repression:
![]()
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
HO’s response to increased
concentration of alpha-factor (Roberts
et.al 2000)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
HO |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
CDC3 |
|
|
establishment of cell
polarity (sensu Saccharomyces)* |
|
structural protein of
cytoskeleton |
|
septin ring (sensu
Saccharomyces)* |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SRC1 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SLY1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
BET3 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SNF5 |
|
|
chromatin modeling |
|
non-specific RNA polymerase
II transcription factor |
|
nucleosome remodeling
complex |
||
|
|
USO1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
LSB5 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
GCD2 |
|
|
protein synthesis initiation |
|
translation initiation
factor |
|
ribosome |
||
|
|
MYO2 |
|
|
establishment of cell
polarity (sensu Saccharomyces)* |
|
microfilament motor |
|
actin cap (sensu
Saccharomyces)* |
||
|
|
YRF1-1 |
|
|
biological_process unknown |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
BAR1 |
|
|
pheromone degradation |
|
aspartic-type endopeptidase |
|
periplasmic space (sensu
Fungi) |
||
|
|
BRN1 |
|
|
mitotic chromosome segregation* |
|
molecular_function unknown |
|
nucleus* |
||
|
|
MAD3 |
|
|
mitotic spindle checkpoint |
|
molecular_function unknown |
|
nucleus |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
PEX17 |
|
|
peroxisome organization and
biogenesis |
|
protein binding |
|
peroxisomal membrane |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
This image was taken from the SGD webpage's Expression Connection database.
• This experiment
uses varying concentrations alpha- factor, which is a Saccharomyces' pheromone,
to reveal resulting expression among genes.
HO is repressed
when exposed to alpha factor. It is repressed most at the 5 nM concentration of
aF. HO expression pattern is similar to
other genes involved in the cell cycle (SNF5, BRN1, MAD3), mating (BAR1) , and
structural proteins(PEX17, MYO2, CDC3).
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
HO’s expression in
response to duration of exposure to alpha-factor (Roberts
et.al 2000)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
HO |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
STD1 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
CNM67 |
|
|
microtubule nucleation |
|
structural protein of
cytoskeleton |
|
spindle pole body |
||
|
|
TFC5 |
|
|
transcription initiation,
from Pol III promoter |
|
RNA polymerase III
transcription factor |
|
transcription factor TFIIIB |
||
|
|
SIT1 |
|
|
transport |
|
not yet annotated |
|
not yet annotated |
||
|
|
YMC2 |
|
|
transport |
|
not yet annotated |
|
not yet annotated |
||
|
|
NGG1 |
|
|
not yet annotated |
|
not yet annotated |
|
SAGA complex |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
RLF2 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
MLP1 |
|
|
protein-nucleus import |
|
molecular_function unknown |
|
nuclear membrane* |
||
|
|
RAD17 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
PDR16 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
MCD1 |
|
|
mitotic chromosome
condensation* |
|
molecular_function unknown |
|
cohesin |
||
|
|
FHL1 |
|
|
rRNA processing* |
|
transcription factor |
|
nucleus |
||
|
|
GRX4 |
|
|
oxidative stress response |
|
glutaredoxin |
|
cellular_component unknown |
||
|
|
STP4 |
|
|
tRNA splicing |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SEN34 |
|
|
tRNA splicing |
|
tRNA-intron endonuclease |
|
nuclear inner membrane* |
||
|
|
CLB6 |
|
|
G1/S transition of mitotic
cell cycle* |
|
cyclin |
|
cellular_component unknown |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
|
|
|
|
|
||
|
|
MIF2 |
|
|
mitosis |
|
not yet annotated |
|
not yet annotated |
This image was taken from the SGD webpage’s Expression Connection database.
• This experiment
uses duration of exposure to alpha-factor to reveal resulting expression among
genes. HO is repressed for the entire duration of alpha-factor exposure. HO’s
expression is similar to genes involved in protein synthesis(FHL1, STP4, SEN34,
TFC5) , cell cycle (MIF2, CLB6, MCD1), metabolism(GRX4), transport (SIT1, YMC2),
and structure (CNM67).
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
HO’s expression
during diauxic shift (DiRisi
et.al 1997)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
HO |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
BIK1 |
|
|
mitotic anaphase B* |
|
microtubule binding |
|
spindle pole body* |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
BUD14 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SNU23 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
VAM6 |
|
|
not yet annotated |
|
vacuolar carboxypeptidase Y |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
DUN1 |
|
|
DNA repair |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
PAU7 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
CDC2 |
|
|
nucleotide-excision repair* |
|
delta DNA polymerase |
|
delta DNA polymerase |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
FIG2 |
|
|
mating (sensu
Saccharomyces) |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
This image was taken from the SGD webpage’s Expression
Connection database.
· HO is initially induced and
then repressed during the cells transition from anaerobic to aerobic
metabolism. HO’s expression is similar to genes involved in mating (FIG2), DNA
repair (CDC2, DUN1), and the cell cycle (BIK1). Many of the genes with this
expression pattern have an unknown function.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
HO’s expression during sporulation (Chu et.al 1998)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
HO |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
not yet annotated |
|
not yet annotated |
||
|
|
NPL6 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
SHU1 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
This image was taken from the SGD webpage’s Expression
Connection database.
· HO is slightly repressed during sporulation. Only five genes
were reported as having similar expression patterns all of which are not yet annotated.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
H0’s expression as
histone is depleted (Wyrick et.al 1999)
|
Orf |
|
Gene |
|
|
|
Process |
|
Function |
|
Component |
|
|
HO |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
BUD9 |
|
|
polar budding |
|
molecular_function unknown |
|
cell |
||
|
|
IMP2' |
|
|
DNA repair* |
|
transcription factor |
|
cellular_component unknown |
||
|
|
PMT2 |
|
|
O-linked glycosylation |
|
dolichyl-phosphate-mannose--protein
mannosyltransferase |
|
endoplasmic reticulum |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
FUN11 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
cellular_component unknown |
||
|
|
LIP5 |
|
|
fatty acid metabolism |
|
not yet annotated |
|
not yet annotated |
||
|
|
CDC24 |
|
|
establishment of cell
polarity (sensu Saccharomyces)* |
|
signal transducer* |
|
not yet annotated |
||
|
|
TAF17 |
|
|
transcription initiation,
from Pol II promoter* |
|
general RNA polymerase II
transcription factor |
|
TFIID complex* |
||
|
|
SUR1 |
|
|
mannose-inositol-P-ceramide
(MIPC) metabolism |
|
not yet annotated |
|
not yet annotated |
||
|
|
RGS2 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
SNF7 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
GPE2 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
TOM70 |
|
|
mitochondrial translocation |
|
protein transporter |
|
mitochondrial outer
membrane translocase complex |
||
|
|
FUN26 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
ERV46 |
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
CBC2 |
|
|
not yet annotated |
|
not yet annotated |
|
not yet annotated |
||
|
|
ELP2 |
|
|
transcription |
|
not yet annotated |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
ECM1 |
|
|
not yet annotated |
|
molecular_function unknown |
|
not yet annotated |
||
|
|
|
|
|
biological_process unknown |
|
molecular_function unknown |
|
not yet annotated |
This image was taken from the SGD webpage’s Expression
Connection database.
· HO is repressed when histone is depleted. HO expression pattern
under this condition is similar to the expression pattern of genes involved in asexual
reproduction (BUD9), DNA repair (IMP2’), metabolism (LIP5, SUR1), signal transduction (CDC24, TAF17), and cell
cycle (ELP2).
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
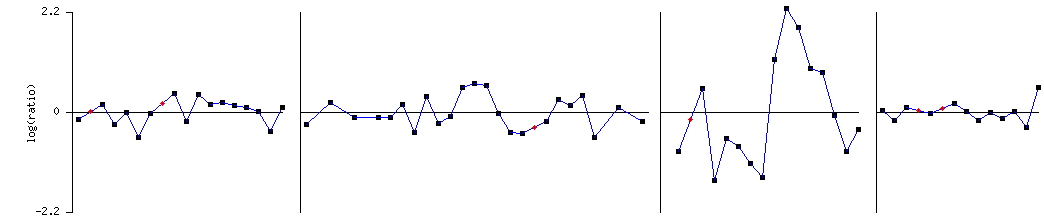
HO’s Expression
during Cell Cycle (Spellman
et.al 1999)
|
Name |
Score |
Peak |
|
|
2.861 |
G1 |
|
|
|
Reference Genes |
|
||
|
10.9 |
G1 |
|
|
|
10.68 |
S |
|
|
|
3.08 |
G2 |
|
|
|
6.726 |
M |
|
|
|
11.8 |
M/G1 |
|
|
|
|
|
|
|
|
Plot of HO (YDL227C) |
|
||
· This experiment analyzes the expression of genes during the cell
cycle. HO is repressed and induced during different stages of the cell but with
a little deviance from 1:1 fold expression.
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾¾
HO’s expression
pattern in the experiments supports the assumption that it is involved in
mating type switch. It is repressed in conditions in which the cell is
committed to a mating type such as sporulation and when exposed to pheromones. The
fact that its expression patterns are similar to DNA repair genes and structural
genes is interesting and surprising in my opinion. Perhaps these genes function
in the processes that HO is involved in when it is induced.
Return to Microarray Main Page