"This web page was produced as an assignment for an undergraduate course at Davidson College."
My Favorite Yeast Proteins
Throughout this semester, my knowledge about the field of Genomics has grown immensely. Initially, I learned how to gather gene sequence information and use databases to gather information and make predictions about the role of the proteins encoded by the genes. Then I learned to analyze DNA microarray data, which provided further information about the molecular functions and biological processes of the chosen genes. DNA microarrays provide information about the particular environments and time points at which genes are induced and repressed. I chose to study two genes from yeast chromosome XVI, FHL1, an annotated gene and YPR098C, a non-annotated gene. Return to my homepage to see the previous two webpages I have created discussing these two genes.
Now I am learning about the field of Proteomics and I have become familiar with new databases that provide protein information. In this last assignment, I hope to uncover some interesting information about the molecular function, biological process, cellular component, and expression of my two chosen proteins. The results that I obtained are below:
My Favorite Annotated Gene
In the previous two assignments I learned that the yeast gene, FHL1, on chromosome XVI, encodes a protein that is a transcription factor. This transcription factor has a fork head domain that regulates transcription of the RNA Pol III promoter, which is involved in in rRNA processing. This protein is not hydrophobic, has many coiled coil domains, is located in the nucleus, and is known to promote terminal development in yeast. The protein sequence of the conserved fork head domain is also observed in this protein's homologs, rat HNF-3, mouse myocyte nuclear factor, and human transcription factor HFK2. In DNA microarray experiments, FHL1 is not induced or repressed in many experiments, but it is repressed under heat shock conditions, high concentrations of hydrogen peroxide and DTT, and during stationary phase experiments. FHL1 is slightly induced in mitosis and kinase A DNA microarray experiments.
Although I found quite a bit of information about FHL1 in my previous searches, I didn't learn much about the interaction of the FHL1 protein with other proteins. I found a paper by Cherel and Thuriaux stating that the protein encoded by the IFH1 gene interacts with the FHL1 protein and changes it from a transcriptional repressor to a transcriptional activator. Interestingly, Cherel and Thuriaux showed that when FHL1 is deleted, yeast grow slowly and have lower amounts of rRNA, but when IFH1 is deleted, the yeast die. They also stated that when both IFH1 and FHL1 are deleted, the yeast cells survive and have normal growth! Therefore, they concluded that IFH1 is essential for growth, but only in the presence of functional FHL1 protein; so these two proteins must interact directly. The Saccharomyces Cerevisi ae Genome Database (SGD webpage) also stated that the FHL1 protein has been shown to interact with transcription initiation factors, DNA binding proteins, and protein tagging proteins, so I was interested to learn more about these protein interactions.
Therefore, I performed searches using various databases to gather information about FHL1 protein interactions. Surprisingly, the first database that I searched, Pathcalling, showed that there were NO known protein interactions with the FHL1 protein. The Interactions Grid Database link from the SGD summary page displayed that the FHL1 protein was identified in association with 5 other proteins. These interaction data were acquired through affinity purification and yeast two hybrid experiments. Of the five proteins found to interact with FHL1, one is a translation initiation factor, three are DNA binding proteins, and one is involved in protein tagging. These results are logical due to FHL1 protein's role as a transcription factor. I would expect that the FHL1 protein would interact with proteins having the described functions in order to carry out it's role in the cell.
*Click here to see the table of interacting proteins from the Interactions Grid Database.
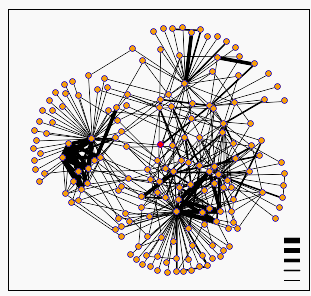
Another database that provides useful information on protein interactions is the Database of Interacting Proteins (DIP) created by David Eisenberg. When I searched this database for the FHL1 protein, I obtained the following graph:

Figure 1: The resulting graph from a Database of Interacting Proteins (DIP) search for proteins that interact with yeast protein FHL1p. The protein encoded by the FHL1 yeast gene is highlighted red and the graph represents all hypothesized protein interactions, with the thickness of the lines representing the probability that the interaction will occur. All of the dots are linked to descriptions of the proteins on the original webpage (DIP, 2003; http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=3821).
When I clicked on the seven closest dots, I found that the proteins predicted to interact with the FHL1 protein were involved in the following processes: resolving four way junctions in mitochondrial DNA, regulating the catalytic activity of casein kinase, which is involved in cell metabolism and differentiation, folding proteins, tRNA splicing, importing ribosomal proteins into the nucleus, binding GTP to terminate the M phase, and regulating transcription and the cell cycle. The names of the proteins with these functions are: CCE1 Yeast, KC2B Yeast, CSBY, Se15 Yeast, IMB4 Yeast, TEM1, CC68 Yeast. Since the protein encoded by the FHL1 gene is a transcription factor that regulates rRNA processing and development, it is logical to me that it might interact with proteins involved in differentiation, tRNA processes, importing ribosomes, and regulating transcription and the cell cycle. And although some of the other protein functions don't seem as relevant, such as resolving four way junctions in mitochondrial DNA, it makes sense that a protein involved in rRNA processing and development might interact with this protein, because processes such as development need energy to continue. And ATP, the main energy source of the cell, is synthesized in the mitochondria. More information can be obtained by clicking on all of the proteins predicted to interact with the FHL1 protein on this graph. The DIP webpage and the the Interactions Grid Database were the two most helpful webpages in my search to find information about the protein interactions of FHL1p.
I obtained the molecular weight (MW), 103.5kD, and the isoelectric point (pI), 5.70, of the FHL1 protein from the MIPS database. This information would be helpful when attempting to locate the protein on a 2D gel electrophoresis experiment. Unfortunately, I was unable to find 2D experiments that included this protein.
The TRIPLES database indicated that when a transposon was inserted into the FHL1 gene at various points, transcription was observed during vegetative and sporulation experiments and was more intense the closer the transposon was inserted to the promoter. This information doesn't really provide any extra information about the FHL1 protein, but is some additional information that I found.
I also searched the Prowl, ExPasy2D, and yeast two hybrid databases and the various PDF files associated with the proteomics chapter in the textbook, but I didn't find any additional information.
Since the molecular function, biological process, and cellular component for the FHL1 protein are already known, the only major information that is missing is that describing protein interactions. From the information that I found, it seems that researchers are beginning to gather more information in this area, so the next step would be to design isotope-coded affinity tag (ICAT) and protein chip experiments to quantify the amount of protein expressed in various conditions.
YPR098C
My Favorite Non-Annotated Gene
In the previous two assignments I learned that the small hypothetical open reading frame (ORF) on chromosome XVI, named YPR098C is predicted to encode a protein with many coils, alpha helices, and transmembrane domains. The MIPS database mentioned above, also predicted that this protein was a transmembrane protein with a pre-cursor molecular weight of 11.6kD and a predicted iso-electric point of 10.67. According to data published on the SGD database, yeast cells are viable when the YPR098C gene is deleted, indicating that this protein may not play as vital of a role as it's neighbor, FHL1. Homolog searches also didn't result in any substantial information, so it is difficult to hypothesize the role of the YPR098C protein using the "guilt by association" method. In the previous assignment, I studied the gene expression patterns of YPR098C in different DNA microarray experiments and concluded that the encoded protein is either involved in responding to stress, because the YPR098C gene was induced in heat shock, irradiation, oxidative stress, and aerobic respiration experiments or ATP synthesis coupled proton transport in the mitochondria, because it clustered with many genes exhibiting this function in many of the microarray experiments.
In order to learn more about the molecular function, biological process, and cellular component of this protein, I searched the TRIPLES, DIP, Prowl, and Expasy2D databases and the PDF files associated with the chapter, but unfortunately these searches resulted in no information!
Therefore, since almost no information is known about this protein, I would first be interested in determining the cellular location of this protein. Once the location of this protein is known, then more hypotheses and experiments can be designed to determine it's molecular function and biological process. In order to determine the location of the YPR098C protein, I would make an antibody to the known sequence, tag the antibody with a flourescent molecule, and then look for the location of the flourescence in the cell under a flourescent microscope. Hopefully, this would provide a clue about the location of the protein. Since my hypothesis is that the protein is located in the mitochondria, I could also find a protocol to isolate the mitochondria from the cell and then run the same antibody experiment. The advantage of this experiment would be that I would be almost 100% positive that the protein was in the mitochondria.
In order to determine protein-protein interactions I would perform both an immunoprecipitation experiment as well as a yeast two hybrid experiment. In an immunoprecipitation experiment, beads coated in antibody for the protein of interest are mixed with the cells and detergent and centrifuged. During this process, the protein of interest and any proteins that are attached to it will precipitate with the beads. Then a solution is added to remove the protein from the beads and an SDS-PAGE gel is run. The identity of the proteins interacting with the protein of interest is determined using molecular weights or possibly sequencing. In the yeast two hybrid experiment, some type of reporter system is set up, such as the production of beta-galactosidase by a transcribed lacZ gene, so that it is obvious to the researcher when the gene is transcribed and when it is not. Then the 'bait' protein, the YPR098C protein, would be fused to the DNA binding bomain of a transcription factor and the 'prey' protein, any other protein, would be fused to the activation domain of the transcription factor. Therefore, using the lacZ reporter system, the cell would only turn blue if the 'bait' and 'prey' proteins interacted. Hopefully, I would gain some protein-protein interaction information from these types of experiments.
Researchers are always striving to learn more about the complex interactions that are occurring in every cell, in every organism, at every moment, but that is a monumentous task. Therefore, they have to prioritize where to spend their time. DNA microarray experiments provide important information about the regulation of different genes, but protein expression and interaction are truly the essential components that need to be understood to better understand the way that cells function. Therefore, experiments using proteomics techniques will be steadily increasing over the next few years. Although I was frustrated that I could not obtain any more information about my unknown gene, YPR098C, and it's encoded protein, I feel that the researchers are trying their best to obtain as much information as they can and therefore, there is probably a reason why there is not more information about this gene. It must be difficult to obtain. I mined all of the current data available for this gene and will look forward to learning more about the techniques that will be used in the future to gather more information about proteins and their interactions. In the future, hopefully more information will be learned about the yeast gene, YPR098C. 2D gels and mass spectrometry experiments can provide useful information about protein expression in different environments, but before environments can be designed, the role of the protein must be further understood.
References:
Cherel I and Thuriaux P. (1995) The IFH1 gene product interacts with a fork head protein in Saccharomyces cerevisiae. Yeast 11(3):261-70.
Send comments, questions, and suggestions to: sahenry@davidson.edu