My Favorite Yeast Gene Expression Page!!!
This site was created for an undergraduate Genomics course at Davidson College
New Data on SSL2!!!
Utilizing SGD's Function Junction database I found one protein known to interact with SSL2. Below is a diagram of this relationship.
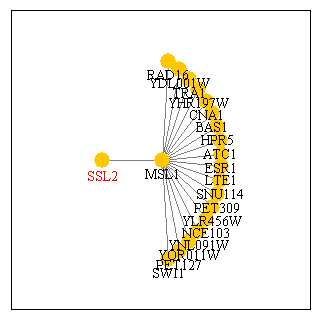
Figure 1: Physical interaction between SSL2 and MSL1. Image provided by SGD Function Junction.
From this figure we see that the only interaction described for SSL2 is with the protein MSL1. MSL1 is an mRNA splicing gene in yeast. We learned from my last web page that SSL2 is involved in nucleotide excision repair of DNA. It is functionally linked with nine other proteins in the TFIIH transcription factor. It appears that this database does not include information about SSL2 relationship with this transcription factor sub-assembly. Furthermore, it is unclear how/why SSL2 would interact with an mRNA splicing protein if it is a DNA helicase. More information on MSL1 is available from SGD here.
Another way to analyze SSL2 is to use DNA Microarrays. This information is available to the public at SGD's Expression Connection webpage. All of the following SSL2 figures were obtained from this webpage. Using this data we can compare what we know and predict about expression of SSL2 to the expression of all other yeast genes under a variety of different experimental conditions. It will be interesting to see if we find other members of the TFIIH showing similar expression profiles to SSL2 since they must all work together to perform nucleotide excision repair.
The following scale is provided by Expression Connection and should be used to interpret the microarray data.

Figure 2: Microarry repression/induction scale.

Figure 3: Expression at different alpha-factor concentrations for SSL2/YIL143C
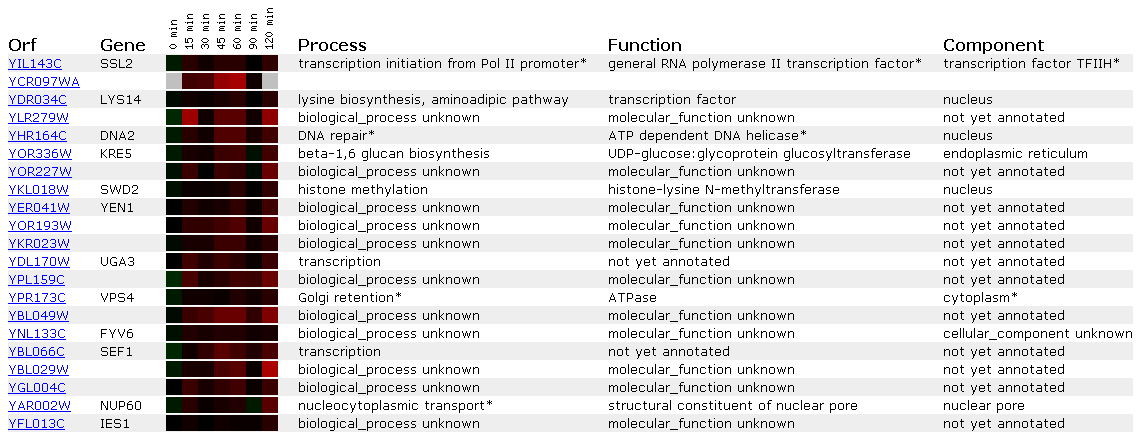
Figure 4: Expression in response to alpha-factor for SSL2/YIL143C
Alpha factor is a pheromone that induces a mating reaction in yeast. Since SSL2 is a DNA repair protein it is most likely not affected by alpha factor. This is shown in Figures 3 and 4 because the SSL2 lane shows no induction/repression compared to the control (all black). Since the expression profile shows no change in response to alpha factor I will not compare the SSL2 to other genes that show similar profiles. This is because a lack of response doesn't strike me as strong enough to infer a 'guilt by association' relationship.
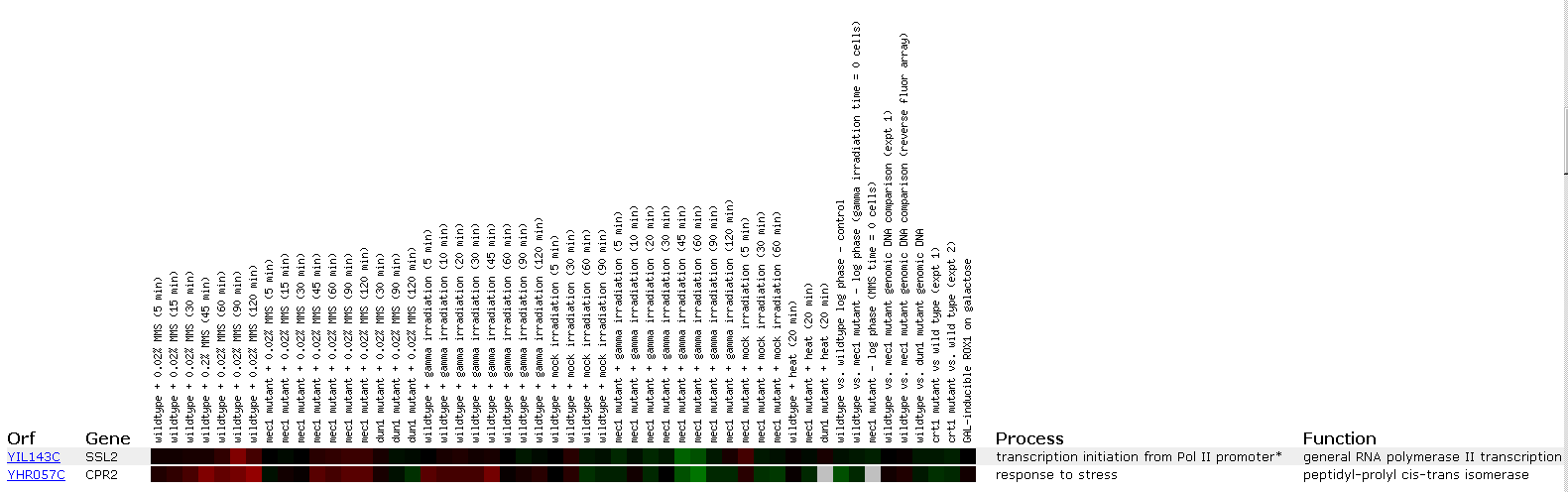
Figure 5: Expression in response to DNA-damaging agents for SSL2/YIL143C
This data is interesting because of the role we know SSL2 to play in DNA repair. We see that at 90 minutes SSL2 is induced when yeast is exposed to DNA damaging agent MMS. It is interesting to note that SSL2 is repressed after 30 minutes of gamma irradiation. This may show that the NEF3 sub-assembly (of which TFIIH is a subunit and SSL2 is a subunit of TFIIH) is not used by the cell in response to this type of DNA damage. The gene CPR2 shows a very similar profile because it responds to stress, and this makes sense because these DNA damaging agents stress the yeast.
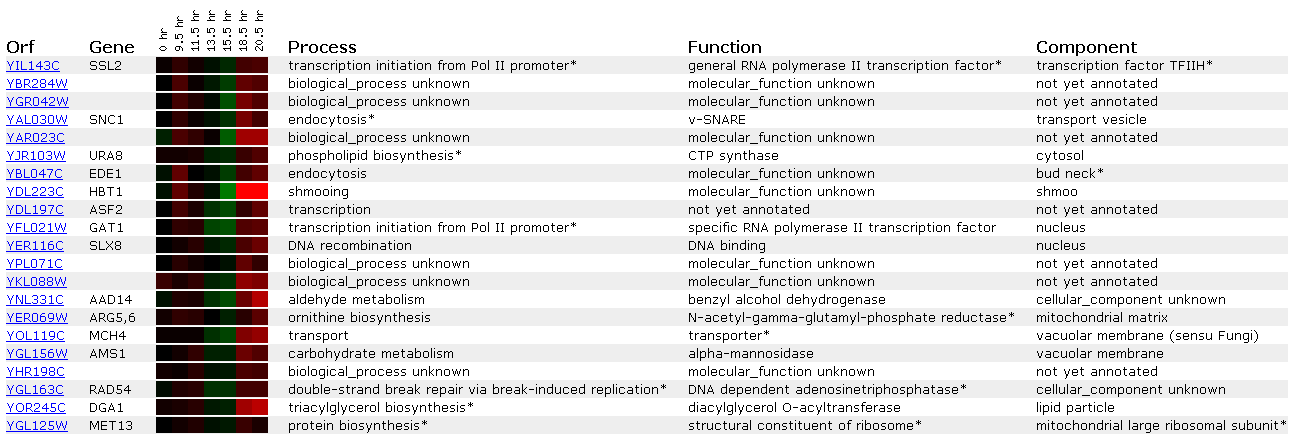
Figure 6: Expression during the diauxic shift for SSL2/YIL143C
A diauxic shift involves changing the yeast growth environment from anaerobic to aerobic conditions. Perhaps this does not lead to any DNA damage because SSL2's is neither induced nor expressed when this event occurs.

Figure 7: Evolution of expression during glucose limitation
This experiment explores changes in transcription between ancestors and decendents after yeast evolve in a glucose limited environment. This data shows no change for SSL2 in the decendants compared to the ancestors, and thus glucose limitation may not play a role in DNA damage.
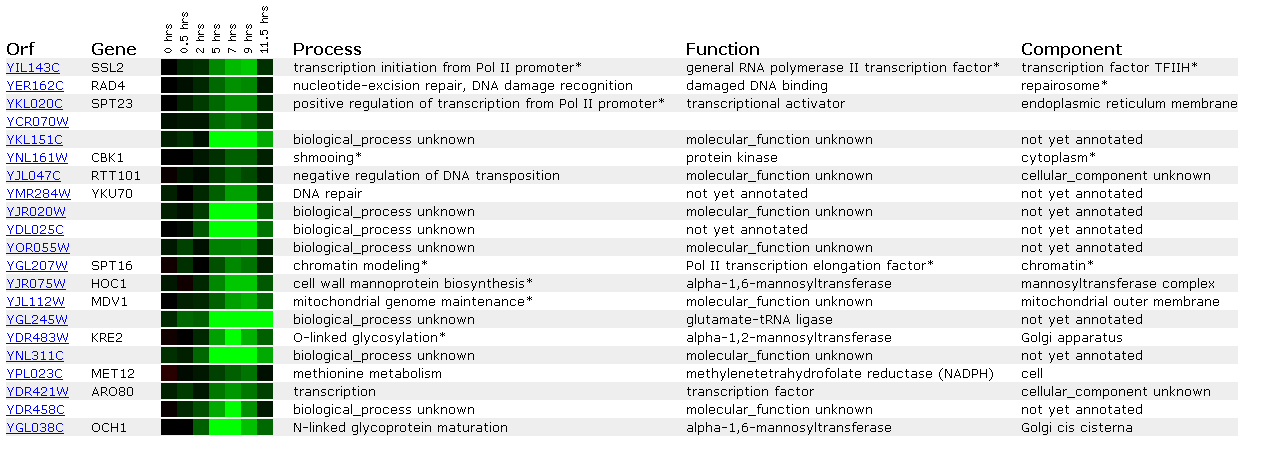
Figure 8: Expression during sporulation for SSL2/YIL143C
Sporulation involves yeast tranforming from diploid into haploid cells and involves meosis. We see from this microarray that many DNA repair genes (SSL2, RAD4, YKU70) are repressed during this process. An explanation for this may be that SPT23, a transcriptional activator for the Pol II promoter, is repressed. When this gene is repressed, SSL2 will be repressed because it is perhaps controlled by SPT23.
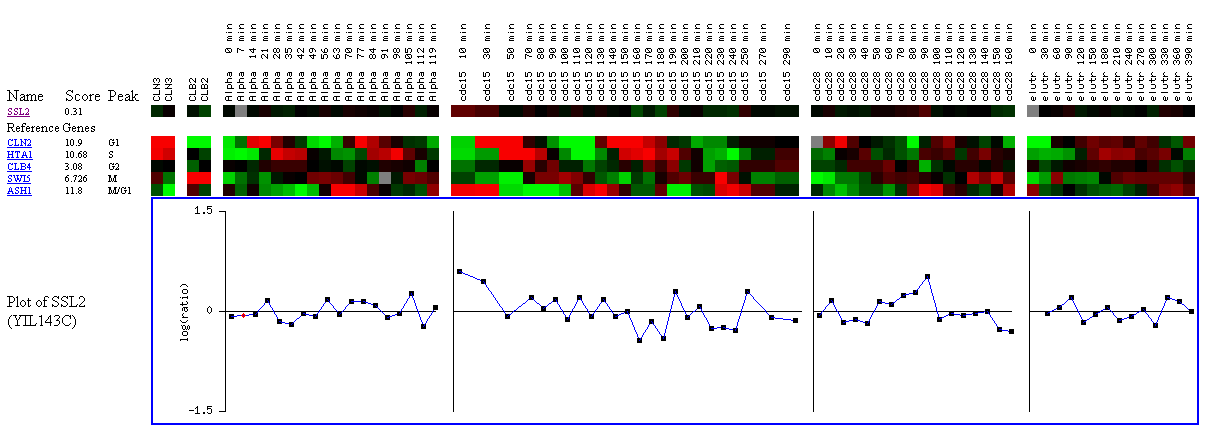
Figure 9: Expression during the cell cycle

Figure 10: Expression in response to environmental changes for SSL2/YIL143C. This is only a portion of the microarray. The top lane is SSL2. The next are YDR26W, SLX8, and CRS5.
We see from this figure that SSL2 is induced during Nitrogen depletion, YPD stationary phase. I know very little about these environmental stresses and will thus leave the interpretation at that.
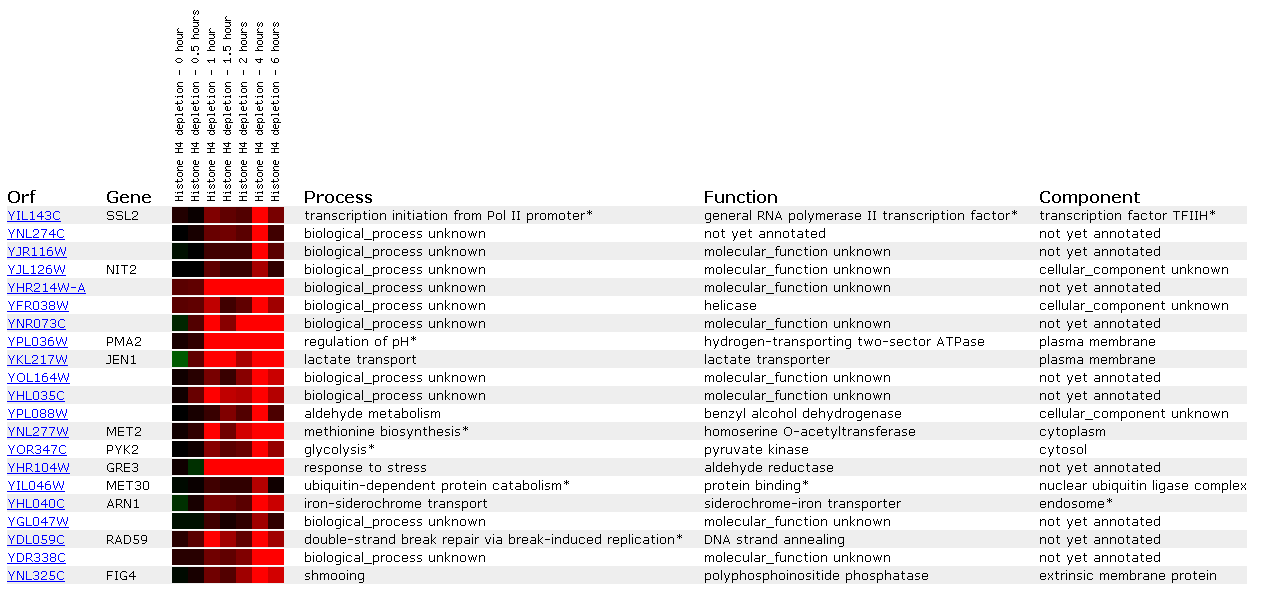
Figure 11: Expression in response to histone depletion for SSL2/YIL143C
This figure shows that histone depletion affects DNA repair genes. Not only is SSL2 induced after 4 hours of depletion, but another helicase (YFR038W) and a double-strand break repair protien that does DNA strang annealing (RAD59) is similarly induced. Since histones are used in winding DNA we may assume that losing histones allows more damage to DNA, and thus induction of DNA repair protiens.
Summary
We have thus used DNA microarrays to explore the expression profile of SSL2 following different experimental treatments. Some figures confirmed our knowledge on SSL2's DNA repair function by showing other DNA repair genes with similar expression profiles. We also learned from the microarrays that showed no change for SSL2. From this data we inferred that DNA damage repair wasn't needed following certain experimental treatments.
New Information on ORF YIL137C!!!
Function Junction: This site provided no information regarding proteins which interact with YIL137C.
Hypothesis after website 2: "From the data presented above we can make predictions about the role of YIL137C in yeast. It seems that it is most likely a leukotriene A4 Hydrolase. Kull et. al. (2001) describe the role of a yeast leukotriene A4 hydrolase, but do not mention its cytogenetic location. The clone they describe seems at least related to this unknown ORF. I hypothesize, based on the domain conservation in this ORF, that YIL137C performs the role of an leukotriene A4 hydrolase which is to "catalyzes the hydrolysis of leukotriene A(4) into the proinflammatory mediator leukotriene B(4)" (Kull et. al. 2001), Another paper about these hydrolases is Haeggestrom, 2000. " (from my 2nd assignment website)
Kull F, Ohlson E, Lind B, Haeggstrom JZ. Saccharomyces cerevisiae
leukotriene A4 hydrolase: formation of leukotriene B4 and identification of
catalytic residues. Biochemistry 2001 Oct 23;40(42):12695-703
Haeggstrom JZ. Structure, function, and regulation of leukotriene A4 hydrolase.
Am J Respir Crit Care Med 2000 Feb;161(2 Pt 2):S25-31
This was the summary about my unknown ORF from web assignment 2. Since then I have changed once aspect of this hypothesis. Reading closer in the Kull paper shows that yeast leukotriene A4 hydrolase does in fact hydrolyze A4 into B4; however, in yeast this is not part of an inflammatory cycle because yeast have no inflammatory response. Leukotriene B4 is only one of 3 compounds leukotriene A4 hydrolase creates. This is an interesting account of a conserved protein performing the same role for two different systems. There is only one amino acid change in the human form that allows the protein to be much for efficient at creating Leukotriene B4 since it plays a new role in human inflammatory response. Check out the abstract of the paper for clarification if needed.
In order to examine whether or not the hypothesized function of YIL137C is in fact true, it is helpful to look at DNA Microarray data. This data is provided, as above, by SGD Expression Connection. The following figures are taken from this site.

Figure 12: Microarry repression/induction scale.
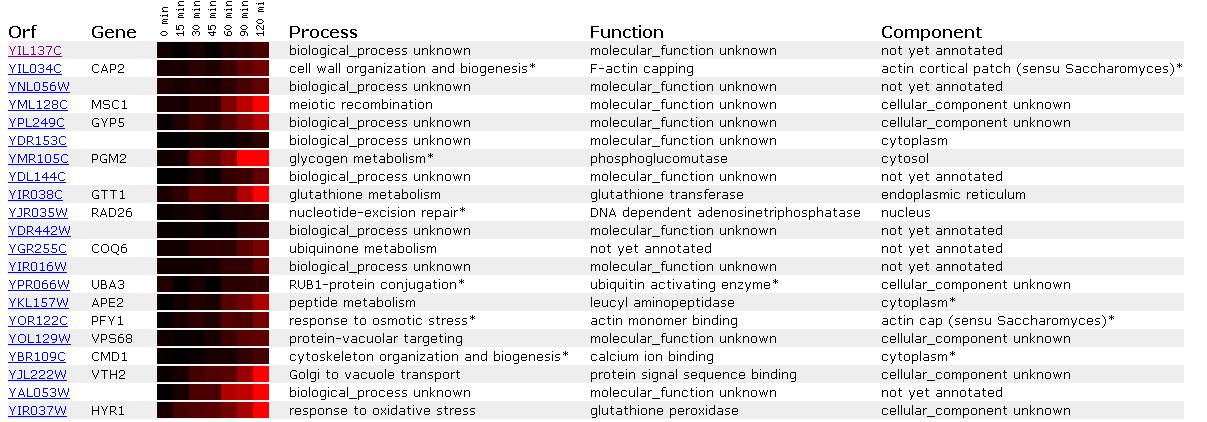
Figure 13: Expression in response to alpha-factor for YIL137C
It appears that this ORF shows no change in gene expression following addition of alpha factor, though it is clustered with genes that are induced. An interesting one to explore is APE2 becuase it is a leucyl aminopeptidase. We recall from web page #2 that YIL137C showed a conserved leucyle aminopeptidase domain. However, since YIL137C is all black and APE2 is induced I am not comfortable using guilt by association to relate the genes.
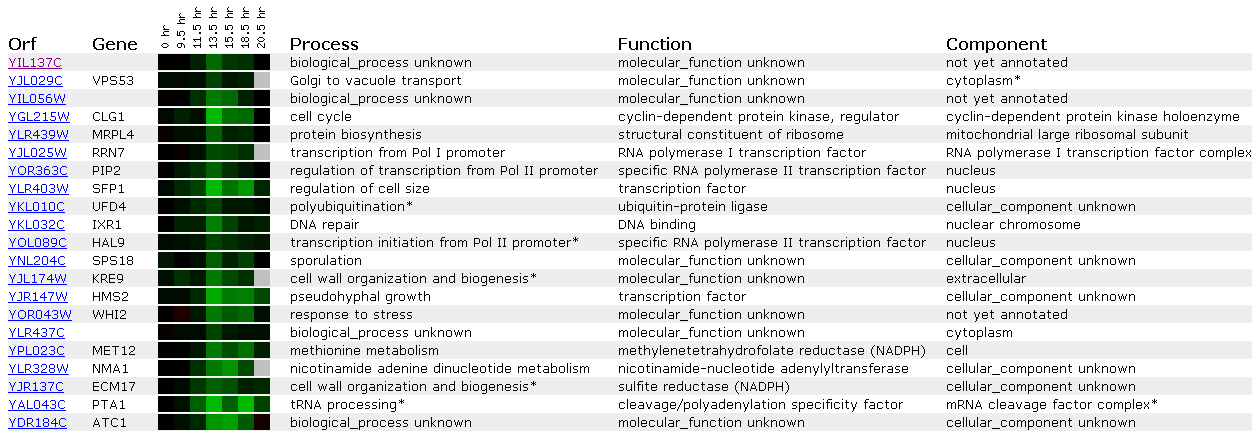
Figure 14: Expression during the diauxic shift for YIL137C
This figure tells us that when yeast are changed from anaerobic to aerobic conditions the YIL137C ORF is repressed. This is the first microarray that shows a similar expression profile betwen this ORF and RNA Polymerase I and II transcription factorrs (PIP2, HAL9, and RRN7). Furthermore we see a DNA repair gene expressed within this ORF (IXR1). SSL2 is involved in a transcription factor for RNA Pol II and performs DNA repair. SSL2 and YIL137C are related only in cytogenetic position, but they may perform similar roles based on guilt by association. Perhaps I was looking for this connection or maybe it is real.

Figure 15: Evolution of expression during glucose limitation
From this figure we see no difference in ancestoral and descendent gene expression profiles following glucose limitation. While it would be more meaningful if there were inductino/repression on this array, I should note that there is a DNA repair gene (CTF4) and am endopeptidase (RPN6). I note these because they are in the categories of the two predicted roles of this unnknown ORF.
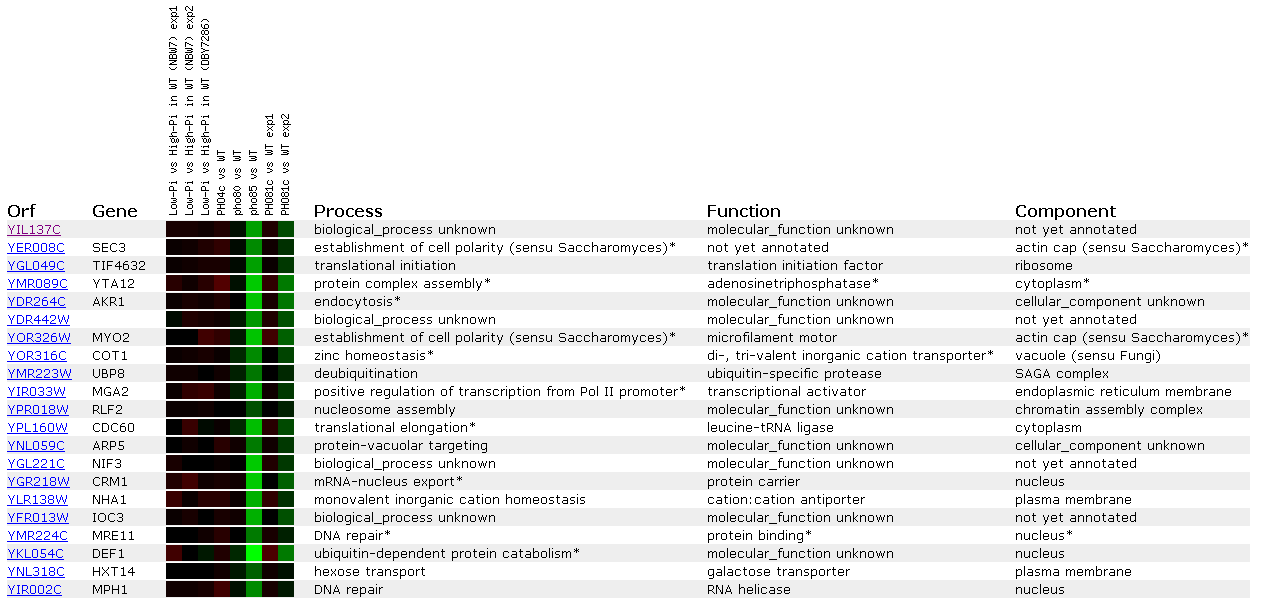
Figure 16: Expression Regulated by the PHO pathway for YIL137C
These data reveal more possible connections between the unknown ORF and DNA repair. Genes MRE11 and MPH1 are both induced similar to YIL137C and both are involved in DNA repair. Also, we see MGA2 that is a regulator of the transcription factors involved with the RNA Pol II promoter.
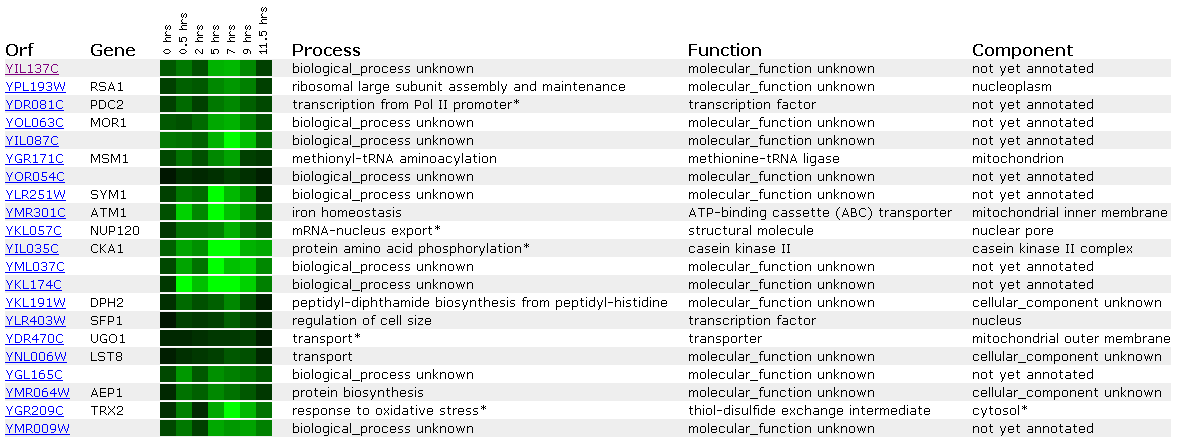
Figure 17: Expression during sporulation for YIL137C
When yeast change from diploid to haploid cells via sporulation, YIL137C is repressed. This is similar to the response seen in SSL2 above. We see in the data PDC2, another transcription factor involved in the RNA Pol II promoter showing a similar expression profile.
Summary
It is not apparent from the microarray data that the hypothesis made on web page #2 is necessarily correct. There was one correlation with a leucyl aminopeptidase in figure 13, however, to my eye the relationship wasn't very strong. Furthermore, I didn't find other expressino profiles for proteins that break down proteins like an aminopeptidase does throughout the arrays. I found the strongest correlation with transcription factors for the RNA Pol II promoter and DNA repair. This is made more intersting by the above exploration of SSL2 since it is involved in this pathway and serves a DNA repair role. I am unsure how often genes that perform similar roles are located near each other on chromosomes, but it may allow for them to be expressed together. However, SSL2 never came up on any of the microarrays and thus I assume that YIL137C and SSL2 are not involved in the exact same aspect of DNA repair. My best assumption at this point is that YIL137C is involved in some sub-assembly that acts as a transcription factor for RNA Pol II and that may (as TFIIH does) serve some DNA repair role.
Contact Kevin James with any comments or questions.