There was no available Chime file depicting the three dimensional structure
of yeast calmodulin. Below is a Chime file for calmodulin in Rattus rattus,
whose calmodulin sequence is 60% identical and 34% similar to yeast calmodulin
(SGD, 2002; <Alignment
of CMD1/YBR109C and PDB 3CLN> Caution: this link will take several minutes
to load.)
Figure 2: 3D structure of rat calmodulin in Chime format. (PDB,
2002; <Structure
Explorer: 3CLN>) Click here
to download the Chime plug-in.
First discovered in 1970, calmodulin is ubiquitously expressed
in vertebrates, where it regulates a number of proteins and processes. According
to Ohya and Botstein, calmodulin interacts with over 20 proteins, including
"several metabolic enzymes, protein kinases, a protein phosphatase, ion
transporters, receptors, motor proteins, and cytoskeletal components"
(1994). Stanford's Saccharmomyces
Genome Database summarizes calmodulin's essential cellular funcions in
terms of the hierarchy set forth by the Gene
Ontology Consortium: biological process, biological function, and cellular
component. Using these categories as a starting point, I have assembled a
brief description of the why, what, and where of calmodulin
in S. cerevisiae
Biological Process:
One process in which calmodulin is involved is mitosis. The
spindle pole body (SPB) is an organelle responsible for the nucleation of
microtubules. This is a function equivalent to that of the centrosome in animal
cells. One component of the SPB is a protein called Spc110p (aka Nuf1p), which
calmodulin binds to at the central plaque of the SPB. This apparently anchors
Spc110p to the spindle pole during mitosis. Calmodulin and Spc110p mutants
both exhibit defective spindle formation and loss of microtuble attachment
to the spindle pole (Cyert, 2001). Calmodulin also performs a role in budding,
by binding Myo2p, a type of myosin that is thought to have play a role in
polarized growth by transporting secretory vesicles to the bud tip. Mutations
in CMD1 and MYO2 both lead to similar defects in bud emergenc. Calmodulin
is also required for endocytosis. It is known to interact with Myo5p and Arc35p.
Yeast carrying the mutant allele cmd1-226 have been shown to display
defects in endocytosis, while not interacting with Myo5p. Interestingly enough,
the previous three functions are not dependent upon calmodulin's ability to
bind Ca2+ (Cyert, 2001). Finally, calmodulin is involved in Ca2+-dependent
signalling pathways that are triggered by stress and environmental changes.
The target proteins for these interactions are calcineurin, a Ca2+ calmodulin-dependent
protein phosphatase, Cmk1p, and Cmk2p, both of which are calmodulin-dependent
protein kinases. Whereas the function of Cmk1p and Cmk2p are not well understood
in terms of signalling pathways, calcineurin is known, in yeast, to be primarily
involved in stress response, regulation of Ca2+ homeostasis, and cell cycle
regulation (Cyert, 2001).
Biological Function:
Calmodulin changes conformation when bound to Ca2+, allowing
it to bind to other target enzymes. Each calmodulin contains 4 structures
called EF-hands, that allow Ca2+ binding to occur. However, yeast calmodulin
can only bind three Ca2+ ions at a time, whereas vertebrate calmodulin can
bind four ions. Although Ca2+ binding is an important function of calmodulin,
calmodulin's essential role in yeast cells is not dependent upon its ability
to bind Ca2+. Yeast cells that have been mutated to express Ca2+-binding-defective
calmodulin show "minimal disruptions in growth and morphology" (Cyert,
2001).
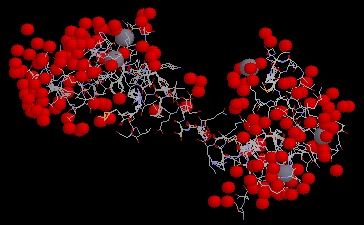
Figure 3: P. tetraurelia calmodulin molecule
shown with bound calcium ions, depicted as grey spheres. The image
is a screen capture taken from a Millenium STING Chime applet. PDB ID = 1EXR
(PDB, 2002; <Structure
Explorer>).
Cellular Components:
SGD lists the locations of calmodulin activity to be incipient
bud sites, the cytoplasm, the central plaque of the SPB, bud tips, bud necks,
and schmoo tips, which is a region of polarized growth (2002).
YBR108W is an ORF located on yeast chromosome 2, near the genes
ALG1 and PHO88. I chose it because of its relative proximity to my favorite
yeast gene, CMD1, as you can see in the map below.
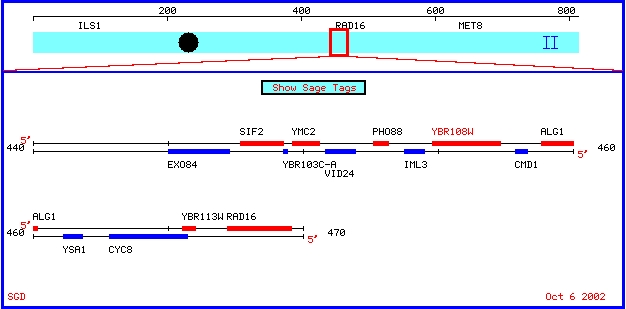
Figure 4: Map showing features around YBR108W on chromosome
2. (SGD, 2002; <Chromosomal
Features Map>).
YBR108W's sequence is 2547 kb long (SGD, 2002; <Sequence
in FASTA format>). Using the Mammalian homolog option at SGD, I retrieved
the following list of alignments, most of which are less than 50% identical
to the portion of YBR108W that they are aligned (2002; <SGD
BLAST Mammal>).
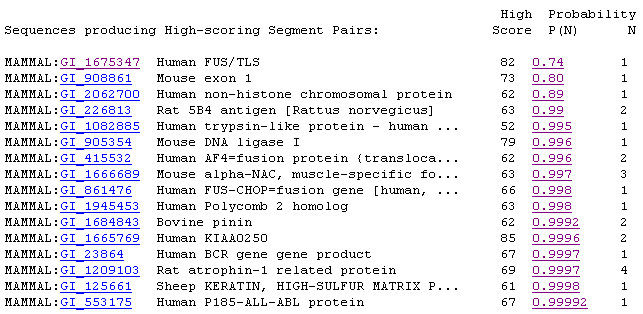
Figure 5: Screen capture of mammalian homolog results.
For the most part, these alignments did not have significant E values
(SGD, 2002; <SGD
BLAST Mammal>).
Using the sequence provided at SGD, I performed a BLASTn
search (Request ID: 1033945276-08698-20824). The two main hits (shown as long
red stripes below, are the NCBI entries for YBR108W). It seems interesting
that several 20-30 kb sequences within YBR108W each produced a number of alignments
(NCBI, 2002; <BLASTn>).
This lead me to wonder if there might be any STS's found within the YBR108W
sequence. However, I did not get any matches using Electronic PCR (NCBI, 2002;
<UniSTS>).
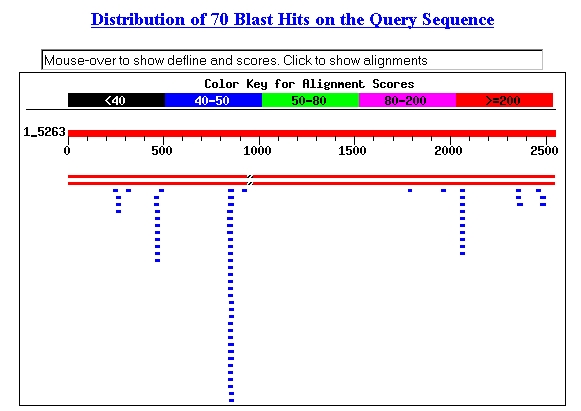
Figure 6: Screen capture of BLASTn hits.
The third line of blue bars from the left had alignments with the following
sequence: acaaccacaacagcaacaacaa (SGD, 2002; <BLASTn>).
SGD predicts the following protein structure for YBR108W (2002;
<YBR108W
Single Page Format>).:
MGFWENNKDSITSGLKSAGKYGYQGTKYVAKTGYKASKKHYNNSKARRER
KSGKKNSSDEEYDSEDEMEYERKPTDIRSLKDPKSFPPPPLKPGQKTYTG
QQQQQMPNGQASYAFQGAYQGQPGAGSTEQSQYAQPQYNQYPQQQLQQGV
MPQQQQLQQGVVPQQPPIYGEQVPPYGSNSNATSYQSLPQQNQPQNAIPS
QVSLNSASQQSTGFVSQNLQYGTQSSNPAPSPSFQNGLQCHQQPQYVSHG
STNLGQSQFPSGQQQQPTTQFGQQVLPSPAQPQQQQQGQPLPPPRGQVIL
PAPGEPLSNGFGQQQQQQQQQQQPLNQNNALLPQMNVEGVSGMAAVQPVY
GQAMSSTTNMQDSNPSYGASPMQGQPPVGGQPPVPVRMQPQPPQPMQQGN
IYPIEPSLDSTGSTPHFEVTPFDPDAPAPKPKIDIPTVDVSSLPPPPTHR
DRGAVVHQEPAPSGKIQPNTTSSAASLPAKHSRTTTADNERNSGNKENDE
STSKSSILGHYDVDANIMPPPKPFRHGLDSVPSEHTTKNAPERAVPILPP
RNNVEPPPPPSRGNFERTESVLSTNAANVQEDPISNFLPPPKPFRHTETK
QNQNSKASPVEMKGEVLPGHPSEEDRNVEPSLVPQSKPQSQSQFRRAHME
TQPIQNFQPPPKPFRRSQSSNSSDSSYTIDGPEANHGRGRGRIAKHHDGD
EYNPKSENSTENGRLGDAPNSFIRKRAPTPPAPSRSEKLHEGTITSEVDS
SKDANKYEKSIPPVTSSIQAQQSTKKAPPPVVKPKPRNFSLKANEYPKEL
TREATGQDEVLNSITNELSHIKLRKTNVNLEKLGGSKKVKTLALFPQI
This sequence contains 848 residues. I performed a BLASTp search
using this sequence, and received the alignment shown in Figure 7. The only
significant hit was the entry for the protein sequence predicted for YBR108W.
The second largest hit was for Zinc metalloprotease in Streptococcus pneumoniae
(NCBI, 2002; <BLASTp>;
Request ID: 1033962783-025846-19786).
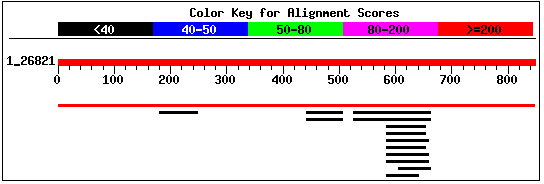
Figure 7: Screen capture of alignments retrieved from
BLASTp search of YBR108W's predicted protein sequence (NCBI, 2002;
<BLASTp>).
I submitted the protein sequence to PREDATOR for prediction
of secondary structure (PBIL, 2002; <PREDATOR
NPS@>). It shows that the predicted structure is over 85% random coil,
and about 10% alpha helix.
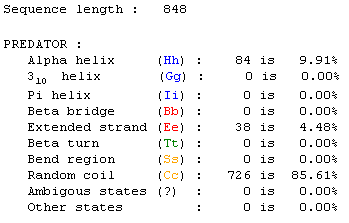
Figure 8: Screen capture received from PREDATOR analysis
of secondary structure (PBIL, 2002; <PREDATOR
NPS@>).
My next step was to enter the predicted amino acid sequence
into a web program that produces a Kyte-Doolittle hydropathy plot. The graph
shown in Figure 9 is the plot that the sequence produced. According to interpretation
information provided on the page, strong negative peaks are an indicator of
a globular protein (Johnson, et al. 2002; <Kyte-Doolittle
Entry Form>).
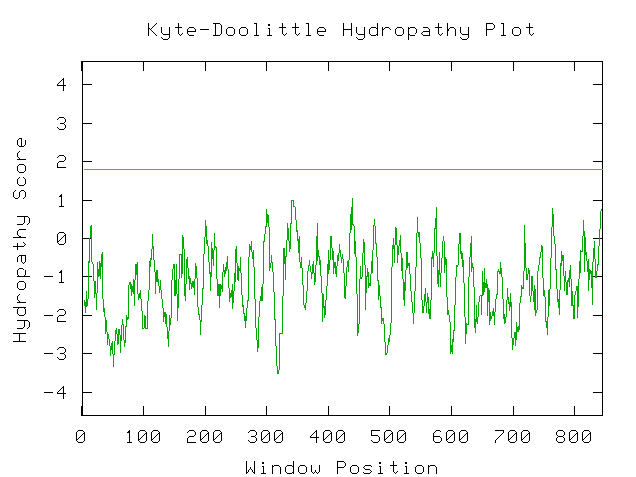
Figure 9: Screen Capture of Kyte-Doolittle Plot produced
from predicted amino acid sequence (Johnson, et al., 2002).
Since my BLAST searches failed to reveal any close matches,
I have chosen Kyte-Doolittle analysis as the source of my speculation as to
the nature of YBR108W. Since the Kyte-Doolittle plot indicated the presence
of a globular protein, I took amino acids sequences for the proteins hemoglobin
(NCBI, 2002; <Hemoglobin
sequence>) and calcineurin b subunit (NCBI, 2002; <Calcineurin
sequence>), which are of known globular structure, and produced Kyte-Doolittle
plots, just to provide bases for comparison. There is a lot of similarity
between the shapes of the Kyte-Doolittle plots of hemoglobin and calcineurin
b and YBR108W's hypothetical protein (Fig. 9). Upon this evidence, I conclude
that the protein which YBR108W encodes is quite possibly a globular protein.
However, at this point I am unable to make any further claims.
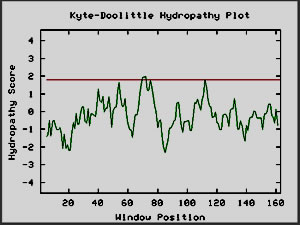
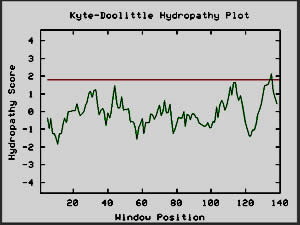
Figure 10: Kyte-Doolittle plots for calcineurin and
hemoglobin I, two globular proteins. The Kyte-Doolittle plot shows
negative peaks, typical of globular proteins (Johnson et al, 2002).
References: