This
web page was produced as an assignment for an undergraduate course at Davidson
College.
My
Favorite Yeast Protein:
AUS1 &
YOR012W
AUS1
SGD:
(SGD, 2002, <http://genome-www.stanford.edu/Saccharomyces/>)
Protein Sequence:
>YOR011W Chr 15
MSISKYFTPVADGSLTFNGANIQFGADAQGESKKSYDAEDSMPNPANQLNDITFQAEAGE
MVLVLGYPTSTLFKTLFHGKTSLSYSPPGSIKFKNNEFKSFSEKCPHQIIYNNEQDVHFP
FLTVEQTIDFALSCKFDIPKGERDQIRNELLREFGLSHVLKTIVGNDFFRGVSGGERKRI
SIIETFIANGSVYLWDNSTKGLDSATALDFLEILRKMAKATRSVNLVRISQASDKIVDKF
DKILMLSDSYQLFYGTVDECLTYFRDTLGIEKDPNDCIIEYLTSILNFQFKNKNLGNLSN
SSSASVLKTATGEVTKYTYNSDFDLYDQWKHSSYYRNIKQQIQGSSIDDSIKEVDPSDVS
PIFNIPLKKQLLFCTKRAFQRSLGDKAYMTAQFISVVIQSLVIGSLFYEIPLTTIGSYSR
GSLTFFSILFFTFLSLADMPIAFQRQPVVKKQSQLHFYTNWVETLSTTVFDYCFKLCLVI
VFSIILYFLAHLQYKAARFFIFLLFLSFYNFCMVSLFALTTLVAPTISVANLFAGILLLA
IAMYASYVIYLKNMHPWFVWIAYLNPAMYAMEAILSNELYNLKLDCSETIVPRGPTYNDV
PFSHKACAWQGATLGNDYVRGRDYLKQGLSYTYHHVWRNFGIIIGFLVFFIACTLFASQY
IKPYFNKDEIERNNSRLTRWLPFLNKKRGTRSSARNDSKYVGIPKSHSVSSSSSSLSAVP
YQISPSNKEMALNDYNEQPITETVETQKHIISWKNINYTVGTKKLINNASGFISSGLTAL
MGESGAGKTTLLNVLSQRVETGVVSGEILIDGHPLTDEDAFKRSIGFVQQQDLHLDLLSV
KESLEISCLLRGDGDRAYLDTVSNLLKLPSDILVADLNPTQRKLLSIGVELVTKPSLLLF
LDEPTSGLDAEAALTIVKFLKQLSLQGQAIFCTIHQPSKSVISHFDNIFLLKRGGECVFF
GPMDDACGYFMSHDNTLVYDKEHDNPADFVIDAVGNSNSSAGKDTAEEALTLNKEAIDWS
ALWESSVEKKLVKKETARLEDDARASGVDYTTSLWKQPSYLQQLALITRRQYICTKRDMT
YVMAKYCLNGGAGLFIGFSFWHIKHNIIGLQDSIFFCFMALCVSSPLINQIQDKALKTKE
VYVAREARSNTYHWTVLLLSQSIIELPLALTSSTLFFVCAFFSCGFNNAGWSAGVFFLNY
MLFAAYYSTLGLWLIYTAPNLQTAAVFVAFIYSFTASFCGVMQPYSLFPTFWKFMYRVSP
YTYFVETFVSILLHNWEIKCDMSEMVPGQPLTGQSCGQFMEAFIEEYGGYLHNKNTFTVC
AYCTYTVGDDFLKNENMSYDHVWRNFGIEWAFVGFNFFAMFAGYYLTYVARIWPKVFKII
TKVIPHRGKKPVQN
This database gave
general information regarding the sequence of the AUS1 protein (See below). You
can click on the link to view the protein sequence.
|
|
Transcript
Translation Calculations
|
|
Codon
Bias
|
0.191
|
|
Codon
Adaptation Index
|
0.196
|
|
equency
of Optimal Codons
|
0.522
|
|
Hydropathicity
of Protein
|
0.019
|
|
Aromaticity
Score
|
0.137
|
|
|
|
TRIPLES:
(TRIPLES,
2002, <http://ygac.med.yale.edu/triples/triples.htm>)
TRIPLES found no data on
AUS1.
The
Grid: (The Grid, 2002, <http://biodata.mshri.on.ca/grid/index.html>)
This database showed that
YOR011W (AUS1) is an ABC transporter and is located in the membrane.
In addition, the database showed that AUS1 is associated with YIR009W.
This protein is involved in mRNA splicing in the snRNP U2. More information can be found by clicking on the links below.
AUS1 was identified in association with the following 1 proteins
DIP
(Database of Interacting Proteins): (DIP, 2001, <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>)
This database show the interaction of AUS1 with YIR009W.
In this database, YIR009W is referred to
as a “hypthetical protein.” This
is different than in The Grid site, where YIR009W is referred to as MSL1.
|
DIP
994N
|
BROWSE LINKS
|
|
Protein: ATP-dependent transport protein homolog YOR011w
|
|
|
|
The graph below shows the
interaction of AUS1 (YOR011W) with other proteins. AUS1 is represented by the red dot, in the lower left hand
side of the diagram. YIR009W is
represented by the dot that is directly in the middle.
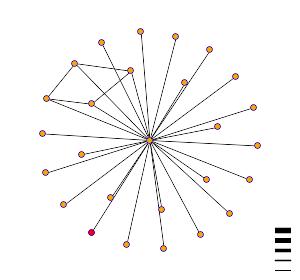
Figure
courtesy of DIP.
MIPS:
(MIPS, 2002. <http://mips.gsf.de/proj/yeast/CYGD/db/index.html>)
This database revealed that
AUS1 is similar to ABC transport proteins.
This is expected because AUS1 is an ABC transport protein.
YRC
Two-Hybrid Analysis &
Additional
Y2H Results:
Found no results for AUS1.
PDF maps:
Aging,
Membrane,
Degration,
Benno
Figure 1:
No result for AUS1.
Function
Junction: (Function Junction, 2002, < http://genome-www4.stanford.edu/cgi-bin/SGD/functionJunction>)
This database revealed that
AUS1 is associated with proteins that serve as ABC transport proteins as well as
proteins that are associated with RNA synthesis. This is similar to the data
found previously.
In addition, Pathcalling presented the
following graph. The graph shows the protein-protein interaction with
regards to AUS1. Once again, you can see that AUS1 (YOR011W) is associated
with MSL1 (YIR009W).
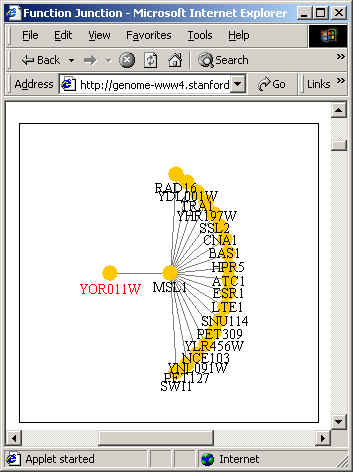
Figure
courtesy of Function Junction
Conclusions:
Although there is not much
information regarding AUS1 in the databases searched, it is apparent that AUS1
indeed codes for a protein that is associated with ABC transport proteins.
In addition, from this search, as well as previous microarray and
literature searches (see website 2 & 3), it seems as though this protein is
associated with replication of DNA in the cell.
YOR012W
SGD:
(SGD, 2002, <http://genome-www.stanford.edu/Saccharomyces/>)
Protein sequence:
>YOR012W Chr 15
MVASSINEESSLAVNLTSDVEKASKTLFKAFEKSYANDYLMKKFFHIPITEKVSRARINA
MIHYYTTCYHDLDGEIAEANDFDAVAIWSRPGCHLPATLSDDESFNKIFFSRLDCEEARS HASGNGLLLPLCHRKRS
In addition, this database
showed the
general characteristics of the protein (See below).
|
Transcript
Translation Calculations
|
|
Codon Bias
|
0.044
|
|
Codon Adaptation
Index
|
0.123
|
|
Frequency of
Optimal Codons
|
0.429
|
|
Hydropathicity of
Protein
|
-0.310
|
|
Aromaticity Score
|
0.102
|
TRIPLES :
(TRIPLES, 2002, <http://ygac.med.yale.edu/triples/triples.htm>)
No data was found for
YOR012W.
The
Grid: (The Grid, 2002, <http://biodata.mshri.on.ca/grid/index.html>)
Returned no data for
YOR012W
DIP:
(Database of Interacting Proteins): (DIP,
2001, <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>)
This database did not
provide the names of any proteins that are similar to YOR012W.
In addition, DIP did not provide a protein interaction map for this
protein.
|
NODE SEARCH RESULTS
|
|
DIP
|
Cross
Reference
|
Protein
Name/Description
|
|
Node
|
Links
|
PIR
|
SWISSPROT
|
GENBANK
|
|
DIP:8948N
|
|
|
Q12351
|
gi:
|
ORF YOR012W
|
MIPS:
(MIPS, 2002. <http://mips.gsf.de/proj/yeast/CYGD/db/index.html>)
This database showed that
YOR012W is related to YDR391C, which is a hypothetical protein.
Since the function of YDR391C is not
known, this provides no information regarding the function of YOR012W.
YRC
Two-Hybrid Analysis & Additional
Y2H Results:
Produced no results.
PDF maps: Aging,
Membrane,
Degration,
Benno
Figure 1:
No result for YOR012W.
No information was gained from Function
Junction.
Yale
Gernstein Lab:
(Yale
Gerstein Lab, 2001, < http://bioinfo.mbb.yale.edu/genome/yeast/search.cgi?orf=YOR012W>)
This database stated that
the protein of YOR012W is located in the cytoplasm.
Possible Experiments:
Although
this page did not reveal any new information regarding YOR012W, the information
that was found on previous pages can be used to hypothesize the function of the
protein. Past pages have suggested that YOR012W is associated with DNA
replication, specifically either with transcription factors or with DNA
repair.
Experiment
1: Proteins involved with
transcription factors or with DNA repair are typically located in the nucleus.
This contradicts the information found by the Yale Gernstein Lab. This lab
found that YOR012W is located in the cytoplasm (see above). Thus, it is
important to test to see where the protein is actually located. One way to
do this would be to use a fluorescently labeled probe. The probe would
contain the complementary sequence of YOR012W, and would move into the
cell via a virus or an injection of antibodies. The probe would bind to
the YOR012W sequence. After binding, scientists could look at the cell
under a microscope and determine where the YOR012W protein is located (in the
cytoplasm or the nucleus) by viewing the labeled probe. Thus, the location
of YOR012W would be determined.
Although
the Gerstein lab believes that YOR012W is found in the cytoplasm, YOR012W
typically clusters with genes associated with transcription and DNA repair.
Thus, it is likely that this protein is associated with these processes and is
found inside the nucleus of the cell. Determining the location of this
protein may lead to a better understanding of its function.
Experiment
2:
The microarray website showed that YOR012W is associated with either
transcription or DNA repair. In order to determine which process it is associated with, an
experiment that involves a modified version of the bar code method by Winzeler
(Campbell & Heyer, 166) could be used. In this method, the YOR012W
gene is deleted and then replaced by a specific marker. This marker will
be easily identifiable, and will function as a sort of bar code later in the
experiment. Other bar coded mutants will also be created.
These mutants will lack proteins that are known to be associated with
replication, such as transcription factors and DNA repair proteins. The
mutants would then be spotted out on a microarray chip.
The
mutants that lacked transcription factors would not replicate and grow.
Eventually, these mutants would die. Mutants that lacked DNA repair
proteins would still grow and replicate, though perhaps at altered rates
(depending on the mutation). By comparing the expression (color) of the
YOR012W mutants to those of transcription factors and DNA repair proteins, it
may be possible to determine in which process YOR012W is involved.
Thus, this experiment may clarify the function of YOR012W.
References:
Campbell,
AM and Heyer, LJ. Genomics, Proteomics, & Bioinformatics
2003. San Francisco, CA: Pearson Education, Inc.
Database
of Interacting Proteins (DIP), 2001, <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>
Accessed 11/11/02
Function
Junction, 2002, < http://genome-www4.stanford.edu/cgi-bin/SGD/functionJunction>
Accessed 11/15/02
MIPS Comprehensive Yeast Genome Database (CYGD),
2002, <http://mips.gsf.de/proj/yeast/CYGD/db/index.html>
Saccharomyces Genome
Database (SGD), 2002, <http://genome-www.stanford.edu/Saccharomyces/>
Accessed 11/13/02
Schwikowski,
Benno; Uetz, Fields. 2000. A
network of protein-protein interactions in yeast. Nature Biotechnology.
18:1257-1261. (PDF files)
The
Grid, 2002, <http://biodata.mshri.on.ca/grid/index.html>.
Accessed 11/14/02
TRIPLES,
2002, <http://ygac.med.yale.edu/triples/triples.htm>.
Accessed 11/11/02
Yale
Gerstein Lab, , 2001, < http://bioinfo.mbb.yale.edu/genome/yeast/search.cgi?orf=YOR012W>
Accessed 11/15/02
YRC
Two-Hybrid Analysis, 2000, <http://depts.washington.edu/%7Eyeastrc/th_11.htm>
Accessed 11/11/02.
YRC
Two-Hybrid Analysis’ Additional YRH Results, 2000, <http://depts.washington.edu/%7Eyeastrc/th_12.htm>
Accessed 11/11/02
Megan's Home Page
Genomics
Web Tools
Genomic
Front Page
Return To Biology Course Materials
Davidson College Biology Department
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: Meshafer@davidson.edu



