This web page was produced as an assignment for an undergraduate course at Davidson College.
Protein-Protein Interaction in Yeast:
Msp1 and YGR031W
Msp1
Msp1 is a mitochondrial protein sorting protein that is located in the outer membrane of the mitochondrion. In my previous websites (my favorite yeast genes and their expression patterns) I used the data from serveral different databases to look at the DNA structures and the expression profile of Msp1 under varying environmental conditions. This data confirmed that Msp1 is a mitochondrial protein embedded in the mitochondrion membrane. In this website I'm going to explore the proteins that interact with Msp1.

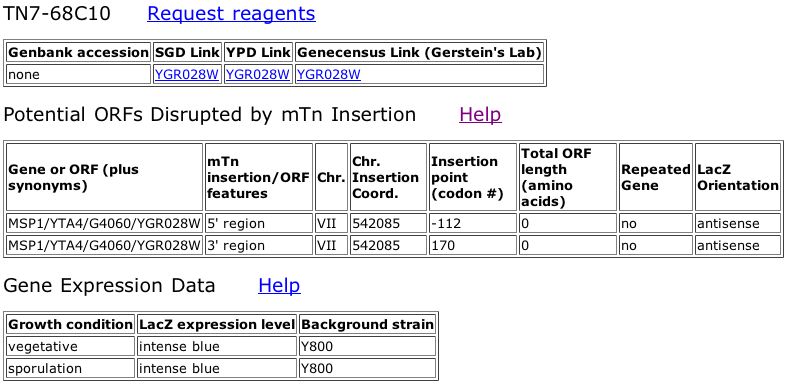
Figure 1. Triples database showing that there is no data for the Msp1 insertion.
The data for Msp1 from the Triples database does not give us any information about Msp1. The insertion of LacZ did not disrupt the gene and there is no phenotypic information given about the mutant.
Pathcalling showed that there were no known interactions of Msp1 with any proteins. I searched PROWL, DIP, Y2H, MIPS (see result in Figure 2) and Enzymes and Metabolic Pathways Database (EMP) and did not find a single piece of information about Msp1 interaction with any protein. In addition, I explored the Swiss-Prot 2D gel database and several other databases of 2D gels and still did not find Msp1. I used the molecular weight and isoelectric point of Msp1 to try to locate them physically on the gels (see Figure 4), but also did searches when possible. Lastly, I explored the pdf files for aging, the Benno Figure 1, degradation proteins, and membrane proteins and did not find Msp1. Since Msp1 is a protein transporting protein, there should be information regarding it's interaction with proteins, although the interaction of Msp1 with the proteins it transports into the mitochondria is probably very fleeting and not easily detected.
Figure 2. MIPS result for Msp1 search indicating that there is no known protein that interacts with Msp1, nor does Msp1 for a larger complex with any proteins.
Figure 3. Pathcalling search results for Msp1 showing that there are no known proteins that interact with Msp1.
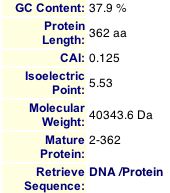
Figure 4. MIPS information for Msp1 showing the molecular weight and isoelectric point which I used to look for Msp1 on 2D gels.
Based on the lack of information regarding the proteins that interact with Msp1, more experiments need to be performed. It would be useful if the Triples database had yielded phenotypic results for the Msp1 insertion mutant because we could have used this information to design more specific tests for Msp1. However, I would use Msp1 as the bait protein in a yeast 2 hybrid system, testing more specifically proteins that have similar expression patterns to Msp1 (see previous website), such as IDH2 and YDC1. I hope to find that several of these proteins interact with Msp1. Since the expression patterns are similar, they are present in the cell at the same times and would therefore be more likely to interact than those proteins that are not similarly expressed. Once we find one or several proteins that interact with Msp1 we can examine the bigger picture because those proteins will likely already be linked to other proteins through interactions and we will have a bigger pool to look at with the yeast two hybrid method.
YGR031W
I had previously asserted that YGR031W is a lypophospholipase that is integral to a membrane, such as the nuclear or ER, and may interact with cytoplasm. In this webpage I aim to further test my hypothesis by looking at protein-protein interactions.
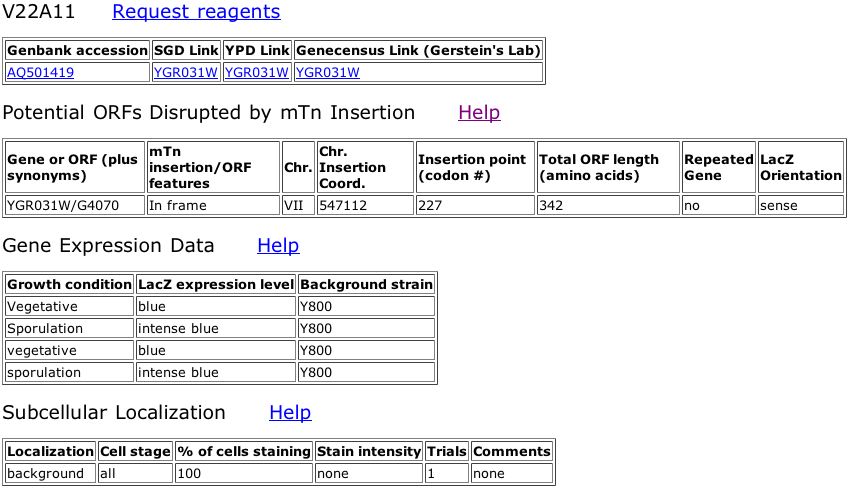
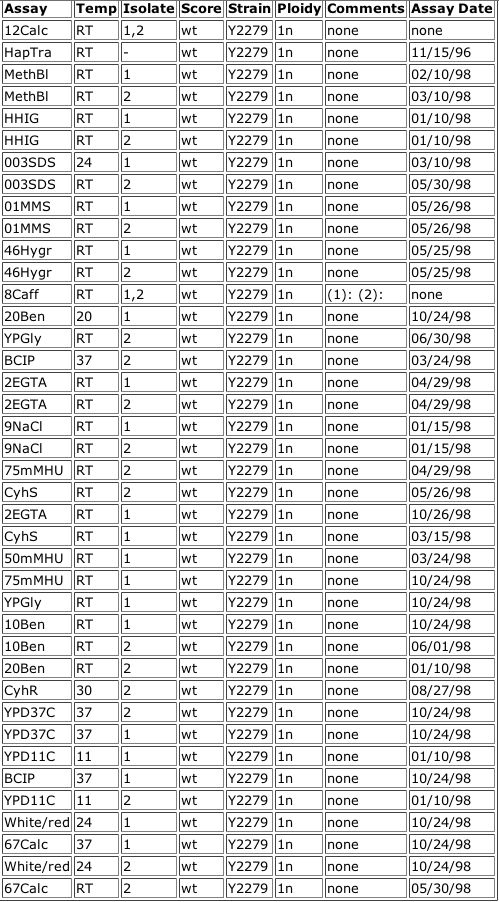

Figure 5. Data from the Triples database showing an inframe insertion of lacZ into YGR031W. The insertion occurs at amino acid 227 out of 324. The subsequent strain of yeast was tested using various reagents in the media to observe any phenotypic changes, but none were observed (all scores were wildtype). A key for the reagents is included.
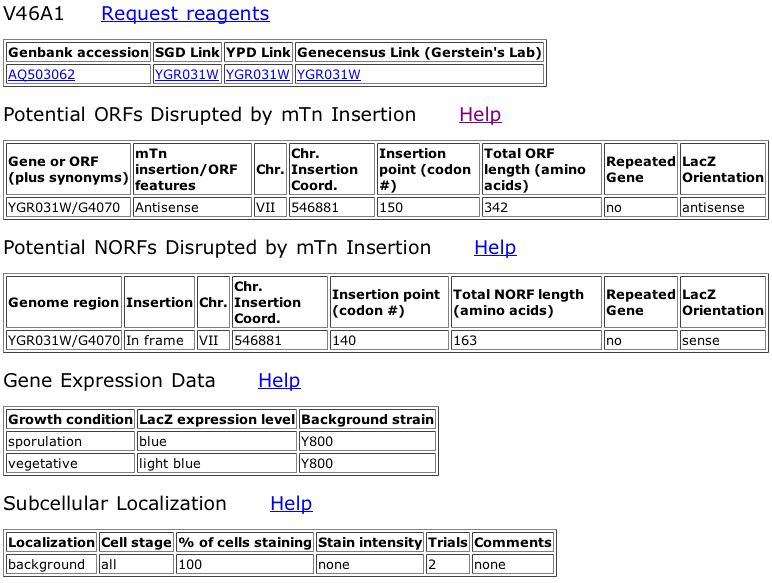
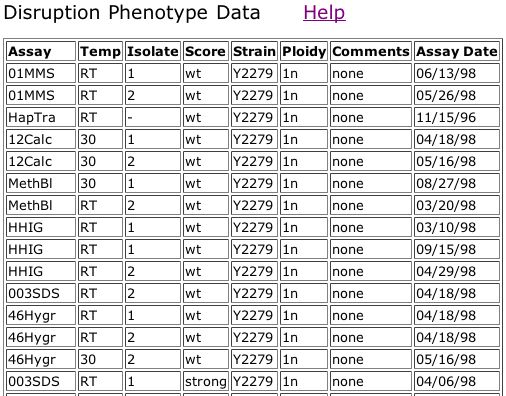
Figure 6. Triples database data for a out-of-frame insertion into YGR031W that shows a strong phenotype when treated with 003SDS (see key in figure 5).
The information from the Triples database shows that the mutant in Figure 6 has a strong phenotype when grown on media containing SDS. The legend from Figure 5 says that mutants demonstrating a phenotype on this media are likely involved in cell wall maintainence or biogenesis. This phenotype does not seem to fit with my prediction that YGR031W is a lypophospholypase since the lypophospholypase is not involved in the cell wall, although it is embedded in a membrane.
I searched PROWL, EMP, and Expasy and did not find any data on protein-protein interactions with YGR031W. I also searched the Swiss-Prot 2D gel database and several other 2D databases linked through Swiss-Prot, but found nothing. The pdf files for aging, Benno Figure 1 (Schwikowski et al 2000), Degradation proteins and Metabolism proteins did not have YGR031 either. However, DIP and MIPS both gave me a direct result (see Figures 7 and 8).
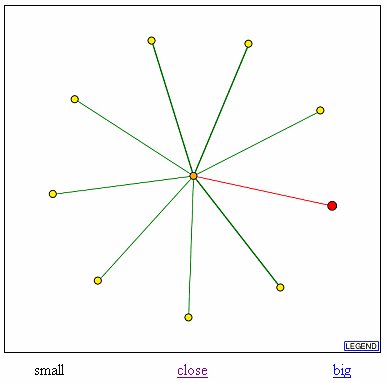
Figure 7. Database of Interacting Proteins (DIP) results for YGR031W showing YGR031W (in red) and it's connection to Bzz1 (center circle). Proteins that also interact with Bzz1 are shown as spokes radiating from Bzz1 in the center.
From DIP, I found that YGR031W interacts with Bzz1. Bzz1 is a yeast protein of yet unknown function. It is has a SH3 homology domain. The SH3 module is thought to mediate the assembly of specific protein complexes by binding to proline-rich peptides. A direct assay has shown that Bzz1 localizes to the actin cortical patch in the cytoplasm. In addition, Bzz1 directly interacts with the myosin heavy chain homolog YKL129c and cytoskeleton assembly control protein SLA1. It's interaction with YGR031W is confusing. If Bzz1 is located in the actin cortical patch, why does it interact with YGR031W? It could be that YGR031W is a lypophospholipase that helps provide some sort of energy to the cell during cell division and it interacts with Bzz1 when the cell undergos stress and needs more energy for the actin/myosin cytoskelton. Or YGR031W could be some other sort of protein that is involved in the actin cytoskeleton. Since the function of Bzz1 is unknown, I cannot draw any definite conclusions for YGR031W, especially since Bzz1 also interacts with elements that are not involved in the actin cytoskeleton such as SEN2, a portion of a nuclear tRNA splicing protein, PSA2, a proteosome, and GTR1, a nuclear phosphate transporter. Therefore, it is very possible that YGR031W is not located in the actin cytoskeleton.
The best way to determine the role of YGR031W is to perform specific experiments that would allow me both to visualize where in the cell YGR031W is located and what YGR031W interacts with. To determine where YGR031W localizes (which would help us determine which proteins to focus on in later experiments in addition to helping us elucidate the function), we could make an antibody to YGR031W would would either fluoresce or have a second antibody that would bind to the YGR031W antibody that would be fluorescent. This antibody could then be used to localize YGR031W within the cell. If I knew where in the cell YGR031W was located, I would be able to choose proteins that are localized to the same region of the cell and expressed at the same time as YGR031W. I would then use these proteins in a yeast two hybrid experiment to test for protein-protein interactions with YGR031W as the bait. Hopefully, the protein-protein interaction will reveal proteins that are already well annotated and the function of YGR031W can be inferred from those interactions. However, if this is not the case, then I could do yeast two hybrid experiments with a wider range of proteins and use both YGR031W as bait and use the proteins that have already been shown to have interactions with YGR031W as bait as well. The point of using the proteins that we found interacted with YGR031W in this second experiment is to hopefully give us clues to their functions, which would in turn help us figure out the function of YGR031W. If all goes according to plan (which it usually doesn't) then we would know some of the proteins that interact with YGR031W and determine it's function and cellular localization.
References
Database for Interacting Proteins (DIP), <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>.
Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proceedings of the National Academy of Science 98(8):4569-74.
Munich Information Center for Protein Sequences (MIPS), <http://mips.gsf.de/genre/proj/yeast/index.jsp>
Pathcalling, <http://portal.curagen.com/pathcalling_portal/index.htm>
Schwikowski et al. 2000. Nature Biotechnology 18:1260.
Questions or comments?: e-mail Sarah Baxley