My Favorite Yeast
Proteins: Exploring Their Roles
My Annotated Yeast Protein
SSD1
(YDR293C)
1. BLASTp
I performed a BLASTp search with SSD1 to find
orthologs and conserved domains. The search returned many results, including
ribonucleases, exonucleases, exoribonucleases and mitotic control proteins (Figure
1). Almost all of the conserved domain hits were included in the exoribonuclease
family (Figure 2). The results suggest that SSD1 may have molecular functions
somehow related to exoribonucleases.
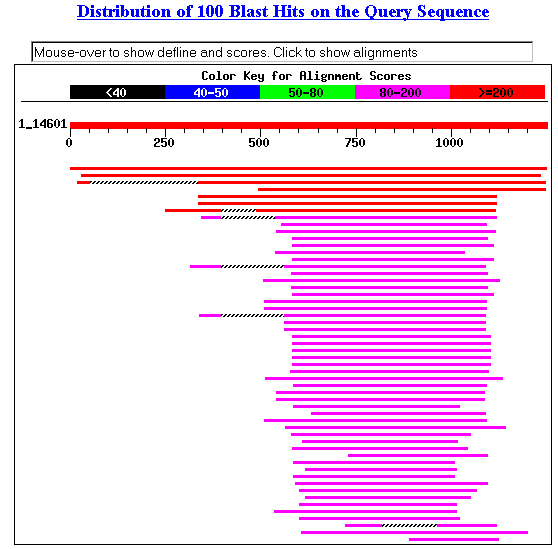

Figure 1. Blastp results with SSD1.
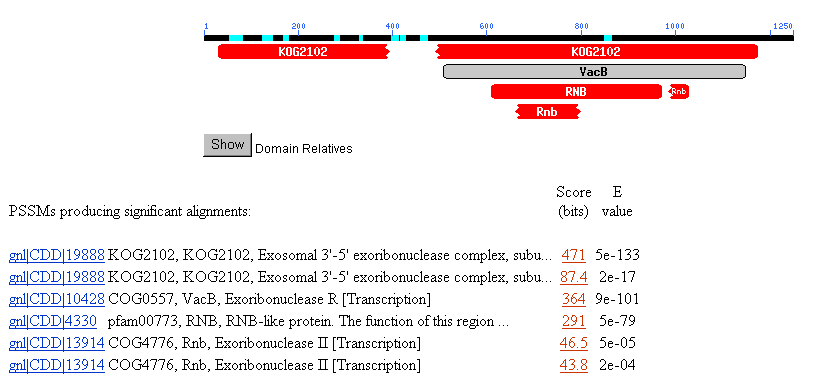
Figure 2. NCBI conserved domain search with SSD1.
2. TRIPLES Database
This database shows results from transposon-insertion
experiments, including the consequences in phenotypes, localization and expression
in yeast. There were five different experiments where the lacZ transposon
inserted itself in SSD1 as reported below. (Clones V108D3 and V178C9 (both KAR2)
were found in chromosome X. My gene of interest, SSD1, is found in chromosome
IV.)
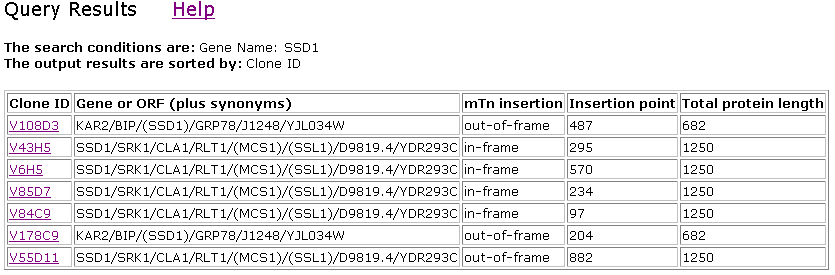
Figure 3. TRIPLES results with SSD1.
Much information was included in each link above. To summarize only the interesting
trends, I noted that there was only one experimental condition that yielded
phenotype disruption (changes in growth compared to wild type) as a result from
the transposon insertion: 46Hygr in Clone V55D11, as shown below in Figure 4.
This striking data reinstates what I have found earlier, that SSD1 protein is
actively involved in cell wall organization and biogenesis.

Figure 4. The only transposon insertion that caused a phenotype change in yeast
growth. From V55D11 insertion.
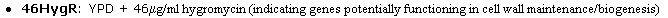

Another interesting information I found was the localization of SSD1 proteins
with transposon insertion.
The following data and picture show where the Clone V6H5 was found.
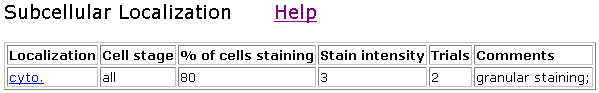
Figure 5. Localization of V6H5. Cyto indicates whole-cell staining.
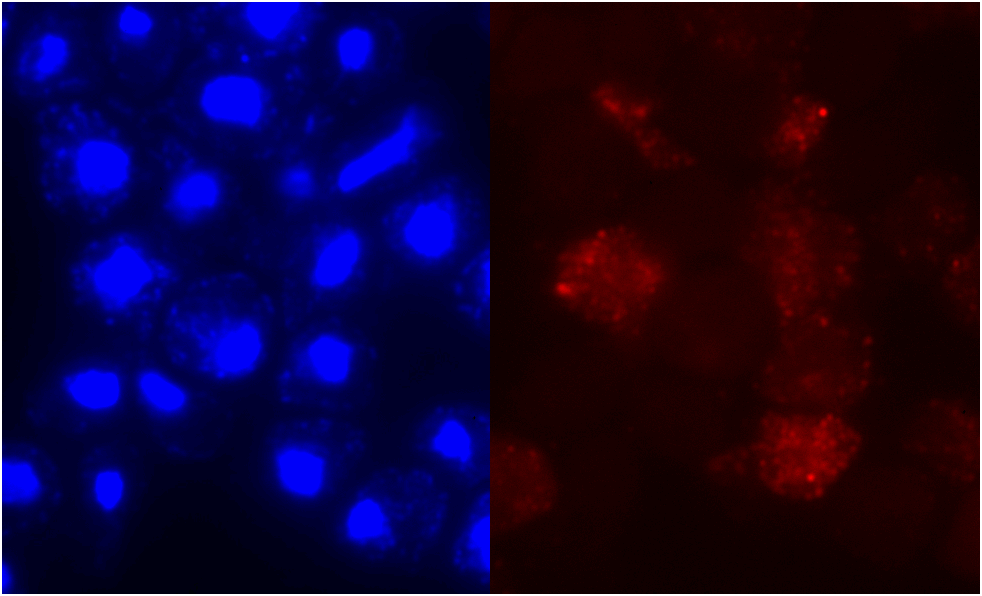 Figure 6. Localization
of clone V6H5. The protein staining was not restricted to any particular cell
stage, and staining intensity was 3 out of 4. The cells were examined by immunofluorescence
with anti-HA antibodies twice for V6H5. The entire cell is stained, although
the nucleus seems to glow brighter.
Figure 6. Localization
of clone V6H5. The protein staining was not restricted to any particular cell
stage, and staining intensity was 3 out of 4. The cells were examined by immunofluorescence
with anti-HA antibodies twice for V6H5. The entire cell is stained, although
the nucleus seems to glow brighter.
Other SSD1 clones showed slightly different staining patterns.
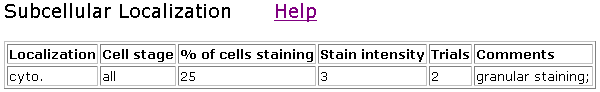
Figure 7. Localization of clone V85D7. This clone showed only about 25% staining.
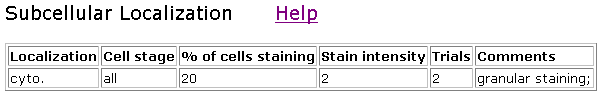
Figure 8. Localization of clone V85C9.
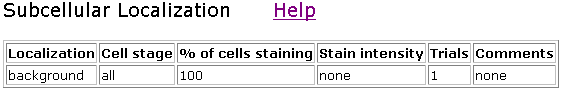
Figure 9. Clones V55D11 and V43H5 did not show any staining above the background
level.
Although the phenotypic change data for V55D11 seemed convincing that SSD1
participated in cell wall organization and biogenesis, I wonder why the rest
of the SSD1 variates did not show the same data. The length of the proteins
were equal at 1250 amino acids. However, it might be that the insertion site
for V55D11 occurred near the end of the protein (at 882 of 1250), while the
other four insertions occurred more upstream (consult Figure 3). This data may
suggest that the amino acids that are upstream of 570 (insertion point of V6H5
which showed no phenotypic change) may be functionally more important.
I was also curious why the localization did not show any preferences in the
different cell stages, because SSD1 has been implicated in G1 cell cycle in
my earlier findings. Perhaps the tansposon insertion somehow interfered with
transcription regulation and prevented the protein from being trascribed at
the right time during the cell cycle.
3. Database of Interacting Proteins (DIP)
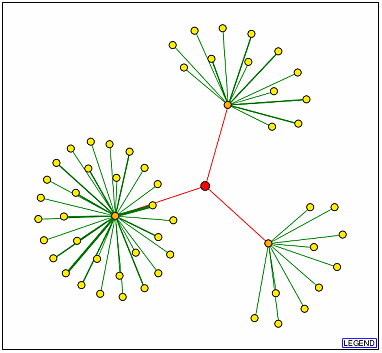
Figure 10. DIP results with SSD1p as the node
(red). The orange dots represent (from the top, clockwise) UTP7p, CBK1p, and
TEM1p.
I have summarized some basic information about the three proteins that are represented
by the orange dots in Figure 10.
The links take you back to the SGD site where the information came from.
| Gene Ontology terms |
UTP7p |
CBK1p |
TEM1p |
| Molecular Function |
snoRNA binding
|
protein kinase activity
|
protein binding
small monomeric GTPase activity
|
| Biological Process |
processing of 20S pre-rRNA
|
cellular morphogenesis during conjugation with cellular fusion
establishment and/or maintenance of cell polarity (sensu Saccharomyces)
regulation of exit from mitosis
response to pheromone during conjugation with cellular fusion
|
M phase of mitotic cell cycle
signal transduction |
| Cellular Component |
small nucleolar ribonucleoprotein complex
|
bud
bud neck
cytoplasm
nucleus
shmoo tip |
spindle pole body
|
It's interesting to note that UTP7 binds to RNA, because my previous findings
from BLASTp conserved domain search indicated that SSD1 may be
functioning as a ribonuclease, so the fact that UTP7 and SSD1 interact with
each other seems to support my hypothesis and the BLASTp results. TEM1 is involved
in M phase of mitotic cell cycle, while SSD1 has been implicated in the G1 cell
cycle.
4. Enzymes and Metabolic Pathways:
I found no results with SSD1.
5. ExPASy 2D Database:
I could only find information about KAR2 (on chromosome X).
6. Benno Figure 1, Aging, Degradation and Membrane PDF
files: SSD1 was not included in the figures.
7. Y2H database: I have
included information from this database in my previous webpage.
The Bottome Line: What
I have found here reinstates my earlier findings, that SSD1p participates in
cell wall organization and biogenesis. Although SSD1p is associated with other
proteins that participate in the cell cycle, according to the TRIPES protein
localization results, there is no significant evidence that SSD1p is found during
a specific stage in cell cycle, suggesting that SSD1p activity is not tightly
cell cycle regulated. The BLASTp conserved domain results seem strikingly consistent
in that all the 7 results had something to do with ribonuclease activity. I
have found earlier that the molecular function of SSD1p was RNA binding and
modification, and thus what I have found in BLASTp seems consistent with my
earlier findings. Six out of the seven CD results suggest that SSD1p contains
a conserved domain with exoribonucleases.
My Non-Annotated Gene
YDR288W
1. BLASTp
I have already performed a BLASTp search with
YDR288W for my previous webpage.
Here, I only show the conserved domain search results.

Figure 11. BLASTp conserved domain results with YDR288W. The two hits show that
YDR288W shares a conserved domain with Melano Antigen, MAGE protein family in
humans. The data suggests that YDR288W may have similar functions as MAGE proteins
in humans.
2. TRIPLES database
This database returned only two results. The
first insertion (Clone TN7-63F11) prevented the protein from being produced,
while the other insertion (Clone V117H1) truncated the protein by about a 100
amino acids. The only positive results were that both insertions tested positive
with b-gal filter assays, turning the cells blue.

Figure 12. TRIPLES result with YDR288W, for Clone TN7-63F11.

Figure 13. TRIPLES result with YDR288W, for Clone V117H1.
3. Database of Interacting Proteins (DIP)
Figure 14. DIP result with YDR288Wp as the node (red). The two orange dots represent
(from the top) QRI2p and QRI1p.
| Gene Ontology Terms |
QRI1 |
QRI2 |
| Molecular Function |
UDP-N-acetylglucosamine diphosphorylase activity
|
molecular function unknown |
| Biological Process |
UDP-N-acetylglucosamine biosynthesis
|
biological process unknown |
| Cellular Component |
cytoplasm
nucleus |
nucleus |
The results are not very helpful, but may suggest that YDR288Wp may be found
in the nucleus or cytoplasm. However, I have found earlier that YDR288Wp is
probably a membrane protein (See my
previous page for YDR288Wp information).
4. Y2H
(YRC Two-Hybrid Analysis)
When QRI1 and QRI2 are used as "Baits", YDR288Wp
was fished out as prey. The results suggest that there are physical protein-protein
interactions between QRI1p and YDR288Wp, and between QRI2p and YDR288Wp. The
physical interaction between the proteins may support my hypothesis that YDR288Wp
may be found in the nucleus or cytoplasm.
| Bait |
Prey |
Prey ORF |
| QRI1 |
YDR288W |
YDR288W |
| QRI2 |
YDR288W |
YDR288W |
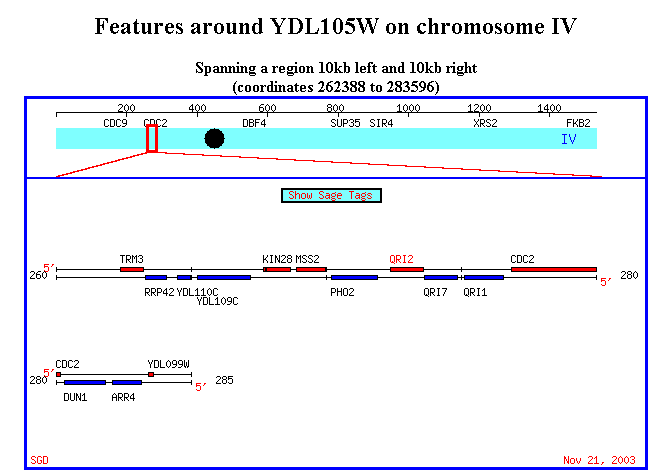
Figure 15. It's interesting to note that QRI1 and QRI2 are placed very close
to each other in chromosome IV. The physical proximity of the two genes may
suggest alternative splicing. Also, it's worth to note that YDR288W is located
in chromosome IV as well, although pretty high upstream of these two genes.
5. ExPASy 2D database:
This database yielded no results with YDR288W.
6. Enzymes
and Metabolic Pathway: no results.
7. Benno Figure
1, Aging, Degradation and Membrane PDF files: YDR288W could
not be found in any of the figures.
The Bottom Line: The
BLASTp conserved domain search suggests more strongly that YDR288W has something
to do with Melanoma Antigen protein found in humans. I have speculated in my
previous webpage, that since MAGE proteins are found in tumors, perhaps
YDR288Wp is involved in cell cycle or cell growth. However, I cannot support
my claim with the information I presented here. MAGE proteins serve as receptors
for T lymphocytes extracted from the tumor-bearing patient; the function of
MAGE proteins seems specifically targeted for the mammalian immune response,
and I am not sure why yeast would produce a MAGE homolog. However, because MAGE
proteins are receptor proteins, this fact supports the hypothesis from the Kyte-Doolittle
hydropathy (from my previous
page) that YDR288Wp is probably a membrane protein. DIP and Y2H databases
show that YDR288Wp interacts with QRI1 and QRI2. The fact that YDR288Wp interacts
with a phosphorylase (QRI1), may suggest that YDR299Wp is post-translationally
modified, such as glycosylation or phosphorylation.
Ideas for Future Experiments:
SSD1 (YDR293C): Continuation of exoribonuclease assay
experiment from Uesono
et al. (method section) in the presence of other proteins that physically
interact with SSD1
My first reaction to the conserved domain search results that related to exoribonucleases
was to test if SSD1 had exoribonuclease activities. However, this study has
already been conducted by Uesono
et al. very thoroughly. However, they have not been able to detect exoribonuclease
activity with SSD1. They discuss several possibilities why their experiment
yielded a negative result: 1) SSD1 may need another component to function properly
as exoribonuclease, 2) formation of a specific complex with RNA may be necessary,
and 3) post-translational modifications may affect the function of the protein
as well. I am not sure why Y2H database did not have any data for SSD1. I would
like to see which other proteins physically interact with SSD1. Using SSD1 as
a bait, I would like to see what the prey proteins would turn out to be. After
determining candidates for proteins that might participate in exoribonuclease
activity with SSD1, I would basically repeat the exoribunuclease assay experiment
that Uesono et al. had conducted earlier. I would expect to see exoribonuclease
activity of SSD1 as a result.
YDR288W: Immunofluorescence labeling with human MAGE
antibodies to locate the cellular component of YDR288W
I would be very interested to see where YDR288Wp localize. With such sequence
similarity to MAGE B-1, I could produce antibodies against the human MAGE B-1
proteins and fluorescently label the antibodies, and let the antibodies bind
to wild type yeast cells. I would like to have YDR288W knock-out yeast cells
as my negative control (this deletion study has already been conducted, and
the cells are still viable after knocking out YDR288W). And human tumor cells
that definitely express MAGE B-1 antigen could work as my positive control.
I reproduce the BLAST2 result from my previous page, because I think antibodies
against MAGE B-1 may work for YDR288Wp with this sequence similarity. I think
locating the protein would be the first step in determining its molecular function
and biological process later on. I am hypothesizing that YDR288Wp is a receptor
protein, so I would expect to see immunofluorescence labeling in the plasma
membrane, which would make the entire cell fluorescent as a result.
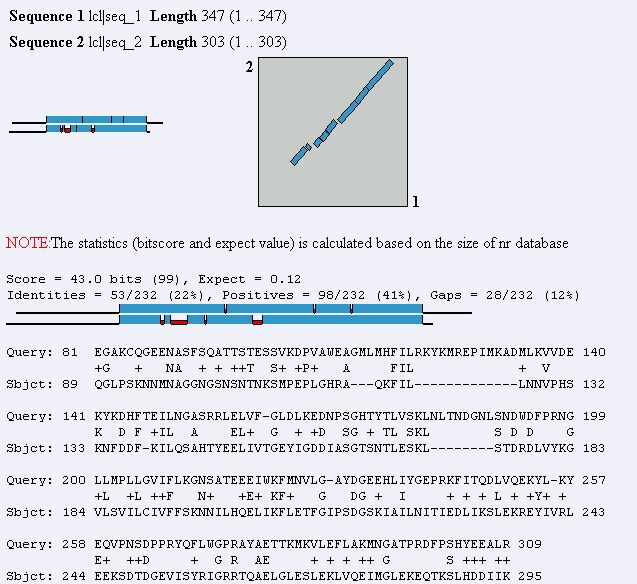
Figure 16. Blast2 result using MAGE B-1 and YDR288W protein sequences. The two
sequences show a fair similarity to each other.
Credits
1. BLASTp and BLAST2 <http://www.ncbi.nlm.nih.gov/BLAST/>
2. TRIPLES <http://ygac.med.yale.edu/triples/>
3. DIP <http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3>
4. Enzymes and Metabolic Pathways <http://www.empproject.com/cgi-bin/map_search.pl>
5. ExPASy 2D database <http://ca.expasy.org/cgi-bin/ch2d-search-ful>
6. Y2H <http://depts.washington.edu/%7Eyeastrc/th_12.htm>
7. SGD <http://db.yeastgenome.org/>
8. Uesono Y, Toh-e A, Kikuchi Y. 1997. Ssd1p of Saccharomyces cerevisiae associates
with RNA. J Biol Chem. 1997 Jun 27;272(26):161039.
<http://www.jbc.org/cgi/content/full/272/26/16103>






 Figure 6. Localization
of clone V6H5. The protein staining was not restricted to any particular cell
stage, and staining intensity was 3 out of 4. The cells were examined by immunofluorescence
with anti-HA antibodies twice for V6H5. The entire cell is stained, although
the nucleus seems to glow brighter.
Figure 6. Localization
of clone V6H5. The protein staining was not restricted to any particular cell
stage, and staining intensity was 3 out of 4. The cells were examined by immunofluorescence
with anti-HA antibodies twice for V6H5. The entire cell is stained, although
the nucleus seems to glow brighter.