Using Proteomic Methods to Elucidate the Putative Roles of Proteins
Introduction
For this discussion I will again treat "my favorite yeast genes," COX5B and YIL103W. Please refer to the two previous investigations concerning these genes (return to inquiries homepage). Herein, I will use several proteomics databases availible online to make an additional conjecture or to revise the earlier hypothesis that I proposed about the role of the protein encoded my unannotated ORF, YIL103W. Finally, I will propose basic research designs to test directly any hypotheses offered in the present discussion.
In general, proteomic research has sought to investigate several experimental questions (Campbell and Heyer 2003). What roles do proteins have in a cell? What is their molecular function or localization? Does a protein's three-dimensional structure reveal its function? With whom does a protein interact in order to execute its cellular role? Which proteins are present in one cell type and not another? Is it possible to quantify the amount of proteins present in a cell? Can allosteric protein modulations be measured? Can answers to these questions be obtained via high-throughput methods, i.e., for more than just a few proteins at a time? Considered together, all of these questions would contribute to the characterization of a given proteome. I shall also begin the analysis of my two yeast proteins with these questions.
Annonated Protein: Cytochrome c Oxidase, chain Vb (Cox5b)
For a more comprehensive account of the current knowledge collected on yeast Cox5b, the reader should refer to my earlier page ("my favorite yeast genes"). I shall, however, provide a brief description of the protein's roles and localizations. Cox5b is one of the thirteen polypeptide chains which comprise the Cytochrome c Oxidase holenzyme (CcO). The entire CcO molecule is responsible for the reduction of dioxygen during oxidative phosphorylation. More specifically, Cox5b and its isoform Cox5a have been shown to both regulate the holenzyme's rate of reaction and are a prerequisite for any redox activity on the part of the CcO. Lastly, Cox5b is induced dramatically during aneorbiosis and is hence known as an anoxic gene in yeast.
SGD database lists the three Gene Ontology classifications for COX5B in the following manner (SGD, 2003; <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=cox5B>):
Molecular Function: cyctochrome c oxidase activity
Biological Process: anerobic metabolism
Cellular Component: mitochondrial membrane, and respiratory chain complex IV
The 3D structure of the CcO holenzyme was isolated and crystalized from bovine heart (Tsukihara, et.al. 1996). In characterizing the whole CcO molecule, each individual chain was also examined. The figure below illustrates the accepted 3D structure.
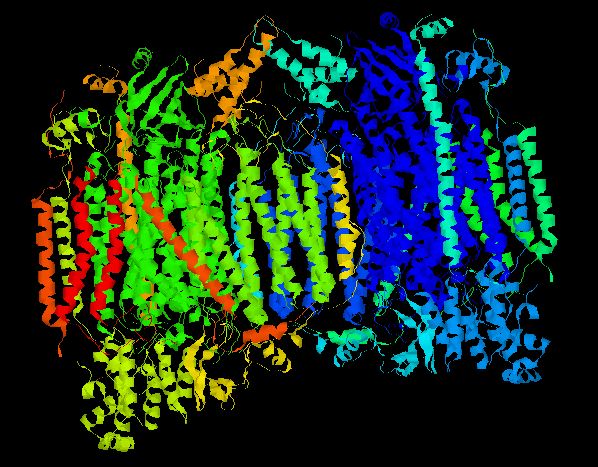
Figure 1 - Crystalized Structure of Bovine Cytochrome c Oxidase. CcO is a dimer composed of two monomers, each possessing all 13 chains. Thus, every CcO molecule has two Vb (Cox5b) chains. Helicies are colored according to their chain number. (PDB, 2003; <http://www.rcsb.org/pdb/cgi/explore.cgi?pid=137651069298962&pdbId=1OCC>).
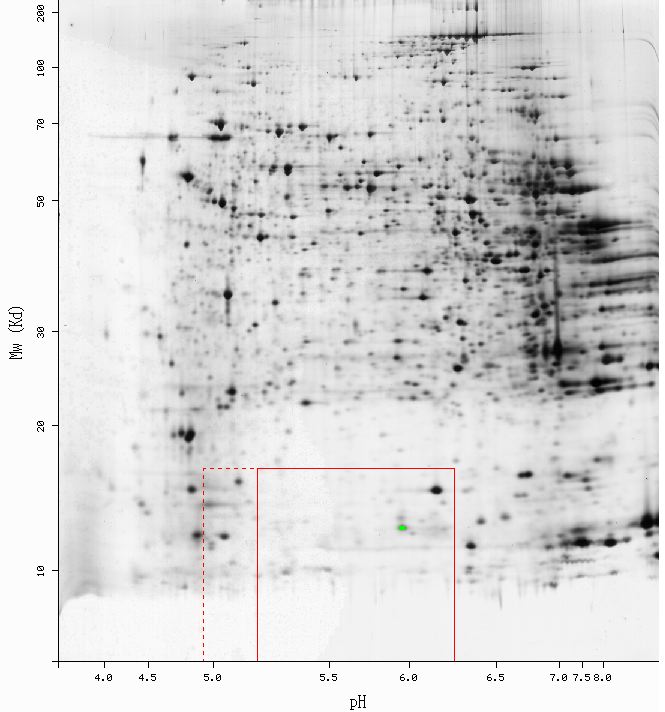
Figure 2 - Results of 2D electrophoresis perfomed using mouse liver tissue. The green point indicates the spot identified as Cox5bp: pI = 5.96, Mw = 12748. No yeast 2D gel data were availible. (SWISS-2DPAGE, 2003; <http://ca.expasy.org/cgi-bin/ch2d-compute-map?LIVER_MOUSE,P19536,2D-0014KV>)
The predicted Molecular weight (Mw) and Isolectric point (pI) for mouse Cox5b were Mw = 13.8 kDa, and pI = 8.69 (ExPASy, 2003; <http://ca.expasy.org/cgi-bin/pi_tool1?P19536@noft@>). As shown in Figure 2, the predicted values are mare larger than the actual values obtaind from the gel. It would be difficult to isolate a single factor responsible for the disparity between the actual and theoretical values, but sample preparation could have introduce extraneous variation. Additionally, it is not suprising that Cox5b was observed in mouse liver tissue. Since the CcO is so vital to oxidative phosporylation, Cox5b is likely to be expressed in the mitochondria of all cell types. Although it is an anoxic gene product, Cox5b has been shown to be active, albeit at very low levels, during aeorbiosis (Burke, et.al. 1997).
With regard to its interaction partners, one can presume that Cox5b interacts extensively with its isoform, Cox5a, for these two isoforms are thought to constitute a synergistic pair, swithing on and off reciprocally over a range of aerobic and anerobic environments (Burke, et.al. 1997). Since Cox5b is thought to be non-functional in isolation, the remaining eleven chains of the CcO holenzyme could be considered interaction partners of Cox5b. Also, Cox5b is reported to be repressed by Rox1p under aerobic conditions (CYGD, 2003; <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YIL111w>). Thus, Rox1p is likely to be another one of Cox5b's interaction partners.
All of the partners just mentioned arise from hypothesis based on the literature. Several online proteomic tools yielded additional results. They are as follows.
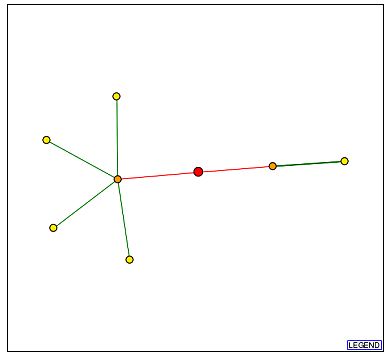
Figure 3a - Schematic graph of the Cox5b interactome. Each point represents a node and is thought to be a protein with which Cox5b interacts. The red point at the center signifies Cox5b, with lines extending to its interaction partners. Beginning with the node closest to the bottom edge and proceeding in a clockwise manner, the yellow nodes are Atp2p, YIR036C, Ayr1p, Atp1p, and Imlp. The orange node to the left of Cox5b is Atp14p, and the orange node to its right is Chl4p. (DIP, 2003; <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=4709>)
Figure 3b - Graph legend for the above schematic of the interactome. (DIP, 2003; <http://dip.doe-mbi.ucla.edu/dip/Main.cgi>)
Although I was unable to retreive any data for Cox5b from the Field's Y2H database, an alternative resource reported bait proteins when Cox5b was the prey. The yeast two-hybrid results depicted immediately below are consistent with two of the nodes in the DIP interactome graph.
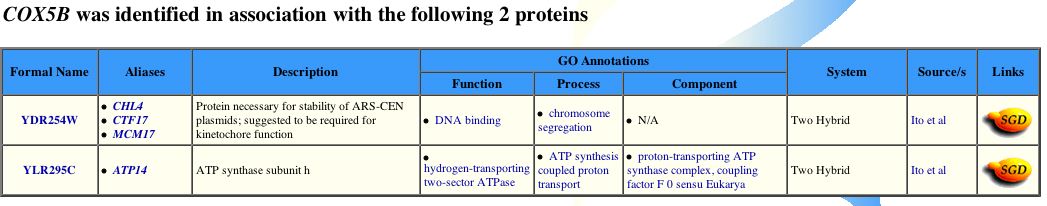
Figure 4 - Positive bait proteins for Cox5b from yeast two-hybrid assays. Cox5b potentially interacts with CHL4 and ATP14. (Yeast GRID, 2003; <http://biodata.mshri.on.ca/grid/servlet/SearchResults?keywords=YIL111W&search=Search>)
In conclusion, proteomic methods have revealed more interaction partners than one would expect from prior knowledge of Cox5b. From both the interactome graph and yeast two-hybrid data, the two partner proteins that warrant most further study are Chl4p and Atp14p. Intuitively, one might expect Cox5b to function inconjuction with Atp14p, which is an ATP synthanse subunit (see Figure 4). ATP synthase spans both mitochondria membranes and is adjacent to the CcO. Moreover, oxidative phosphorylation couples the energy from the oxidation of glucose to transport hydrogen ions, or protons, accross the inner membrane of the mitochondrion. Thus, the translocation of protons into the inter-membrane matrix maintains the proper gradient needed for ATP biosynthesis (Babcok and Wikstrom, 1992). ATP synthase is the holenzyme responsible for catalyzing the reactions necessary for ATP biosynthesis. Their cellular proximity and shared biological roles, however, does not fully explain, a priori, why these two proteins would interact. Both the CcO and ATP synthase holenzymes are fixed in the membrane. Perhaps both Cox5b and Atp14p share a post-translational tag which directs them to the inner mitochondrial membrane upon completion of polypeptide synthesis, and thus they might interact en route to the membrane. On the other hand, Chl4p is more perplexing. The yeast gene, CHL4, YDR254W, encodes the protein Chl4p (see Figure 4; for additional information the reader is refered to Swiss-Prot; <http://us.expasy.org/cgi-bin/niceprot.pl?P38907>). Chl4p appers to assist in the pulling apart of chromosomes during cellular division. The role Cox5b might have during this phase of the cell cycle is unclear.
Unannotated Protein: 103p (YIL103W)*
*I shall remain consistent with my earlier terminology, and for the sake for the present discussion, as before, I shall hereafter refer to YIL103W’s hypothetical protein product as 103p.
It was difficult to uncover any meaningful data for 103p. The fact that the protein is putative explains the lack of data, no doubt. Neither the TRIPLES nor the Field's lab Y2H databases returned any information on 103p. Also, ExPASy 2D gel data were not provided for 103p. The predicted molecular weight and isolectric point were Mw = 48.3 kDa and pI = 8.7 for YIL103W (PROWL, 2003; <http://129.85.19.192/prowl-cgi/sequence.exe/sequence.exe/gi%7C6322088%7Cref%7CNP_012163.1%7C.fsa>). Admittedly, these numbers are of little use on their own, but I have included them should new data become availible in the future.
Despite these setbacks, I was able to locate data which suggest that 103p might have interaction partners in yeast cells.
Figure 5a - Schematic graph of the 103p interactome. The red node at the center represents 103p, and the sole orange node is Dph2p. It should be stressed that a red line indicates that the depicted interaction is unverified. (DIP, 2003; <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=5653>)
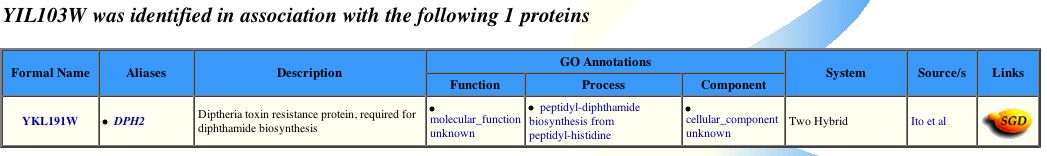
Figure 6 - Positive bait protein for 103p from a yeast two-hybrid assay (Yeast GRID, 2003; <http://biodata.mshri.on.ca:80/grid/servlet/SearchResults?keywords=YIL103W&search=Search>).
In conlcusion, 103p is likely to interact with Dph2p. My conclusion here supports my earlier hypothesis concerning what YIL103W might do in yeast cells. Orginally I proposed that 103p could be "a diphthamide related protein, or might share some of its functional characteristics but have a distinct cellular localization. Upon a consideration of the Conserved Domain data alone, this implication would certainly appear to be true. Although the the BLAST2Seq findings reinforce potential functional similarity between 103p and diphthamide, the low percent identity values, 23% and 21%, with diphthamide proteins precludes the possiblity that 103p and Dph2 are the same protein or even isoforms of the same family" (see page entitled, my favorite yeast genes).
Diphtheria toxin resistence protein 2 (Dph2p) is encoded by
DPH2 (YKL191W) in S. cerevisiae and is thought to be required for
biosynthesis of diphthatmide, which is a modified sturcture of the amino acid
histidine - unique to proteins of the EF-2 translation elongation factor family
(Swiss-Prot, 2003; <http://us.expasy.org/cgi-bin/niceprot.pl?P32461>).
More particularly, Dph2p participates in the “post-translational modification
of peptidyl-histidine to 2'-(3-carboxamido-3-(trimethylammonio)propyl)-L-histidine”
(QuickGO, “GO Term: GO:0017183,” 2003; <http://www.ebi.ac.uk/ego/DisplayGoTerm?id=GO:0017183>).
It is this last reaction that produces diphthamide from histidine.
Diphtheria toxin (DT) has been studied extensively over the past one hundred years. It was of immense biological interest due to the fact that the disease, diphtheria, was a major public health concern. Prior to the advent of vaccinations thereunto, diphtheira was a leading cause of death in infants worldwide. DT is produced by Corynebacterium diphtheirae, which is the bacterium responsible for pathogenesis of diphtheria. DT has become the model protein for the class of bacteiral cytotoxic toxins, which unlike other toxins, terminate its host cell (Stephen 1998). Additionaly, DT belongs to the family of ADP-ribosyl transferases. In its case, DT catalyzes the addition of an ADP-ribose molecule to an EF2p translation elongation factor. Specifically, the diphthamide in EF2p is ADP-ribosylated and as a result, EF2p can no longer translocate a nascent peptide. In this way, DT disrupts protein synthesis, via inhibition of elongation, and the host cell dies. Lastly, DT is composed of two fragments, A and B. The B fragment recognizes certain surface proteins and allows binding to the host cell. Upon binding to the membrane, the A fragment is translocated into the cytosol (Stephen 1998).
Future Manipulative Designs
Furture experiments would need to investigate at least several quesitons. Although it is presemed to reside in the cytoplasm, the precise localization of 103p remains unclear. Additionally, under what conditions is YIL103W expressed? Are any of its transcripts detectable in cells in vitro? Do 103p and Dph2 still interact when 103p is the bait and Dph2p is the prey? Can the orignial yeast two-hybrid data be reproduced? Can 103p be co-immunoprecipated with Dph2 or diphthine synthase? Given its similarity to Dph2p (a protein that functions in production of elongation factors) is 103p likely to be constituative, inasmuch as peptide synthesis is necessary for homeostasis? Can its predicted molecular weight and isolectric point be used to resolve 103p on a 2D gel? Is 103p functional in DPH2 null mutants? Do yeast cells that lack Dph2p and not 103p remain resistant to diptheria toxin? What is the phenotype of a 103p null mutant? Can diphthamide biosynthesis take place in the absence of 103p?
References
Babcock, G.T., and Wikstrom, M. (1992) Oxygen activation and the conservation of energy in cell respiration. Nature 356: 301-309.
Burke, P.V., Raitt, D.C., Allen, L.A., Kellogg, E.A., and Poyton, R.O. (1997) Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J Biol Chem 272: 14705-14712.
Campbell, AM, and Heyer, LJ. 2003. Discovering Genomics, Proteomics, and Bioinformatics. Pearson Education, p.162-194.
CYGD, Comprehensive Yeast Genome Database, "COX5B/YIL111w," Accessed 19 November 2003; <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YIL111w>.
ExPASy database, "Compute pI/Mw [COXB_mouse P19536]," Accessed 19 November 2003; <http://ca.expasy.org/cgi-bin/pi_tool1?P19536@noft@>.
DIP, Database of Interacting Proteins, "DIP: 4709N [COX5B]," Accessed 19 November 2003; <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=4709>.
DIP, Databease of Interacting Proteins, "DIP: 5653N [YIK3_Yeast]," Accessed 19 November 2003; <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?PK=5653>.
DIP, Database of Interacting Proteins, "Graph Legend," Accesssed 20 November 2003; <http://dip.doe-mbi.ucla.edu/dip/Main.cgi>.
PDB, Protein Databank, "Structure Explorer - 1OCC [Bovine Cytochrome c Oxidase]," Accessed 19 November 2003; <http://www.rcsb.org/pdb/cgi/explore.cgi?pid=137651069298962&pdbId=1OCC>.
QuickGO, European Bioinformatics Institute, “GO Term: GO:0017183,” Accessed 21 2003; <http://www.ebi.ac.uk/ego/DisplayGoTerm?id=GO:0017183>.
SGD, Saccharomyces Genome Database, "COX5B/YIL111W - Locus Page," Accessed 20 November 2003; <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=cox5B>.
Stephen, J. 1998. “Toxins,” in Encyclopedia of Immunology, Delves, P.J., and Roitt, I.M., eds. Second edition. Academic Press, 2369-72.
SWISS-2DPAGE database, "P19536 [COXB_mouse]," Accessed 20 November 2003; <http://ca.expasy.org/cgi-bin/ch2d-compute-map?LIVER_MOUSE,P19536,2D-0014KV>.
Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R., and Yoshikawa, S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 angstroms. Science 272: 1136-1344.
Yeast GRID, "COX5B," Accessed 19 November 2003; <http://biodata.mshri.on.ca/grid/servlet/SearchResults?keywords=YIL111W&search=Search>.
Yeast GRID, "YIL103W," Accessed 19 November 2003; <http://biodata.mshri.on.ca:80/grid/servlet/SearchResults?keywords=YIL103W&search=Search>.
Return to Arthur Clement's "Inquiries" Homepage
Any comments, replies, or suggestions would be welcome via email to A. Clement.
© Copyright 2003 Department of Biology, Davidson College, Davidson, NC 28035.
Created on 21 November 2003, by A. Clement.