This web page was produced as an assignment for an undergraduate
course at Davidson College.
Proteomic Data for GPA1 and YHR003C
On two previous websites, I have described the known Saccharomyces cerevisiae gene GPA1 and analyzed information about its non-annotated neighbor YHR003C. One webpage investigated the genomic data to describe the function of GPA1 and to make initial predictions about YHR003C. The second webpage investigated the expression patterns of these genes. Further information about the cellular roles of these two proteins can be deduced from an analysis of proteomic data: information dealing with protein structure, function, and location.
General Proteomic Data Analysis Information
Though analysis of the genetic sequence and expression patterns of a gene may provide important information about the function of the product of this gene, the functional unit in the cell is the protein. Thus, investigations of the structure, function, interactions, and location of the protein product of a particular gene is vital. This "proteomics" analysis is, in general, not as developed as genomic or transcriptomic analysis, but several databases are available to investigate aspects of protein function.
The proteomic data reviewed on this page can be found at the following sources:
TRIPLES- This database of TRansposon-Insertion Phenotypes, Localization, and Expression in Saccharomyces gives information from Yale University experiments in which transposable elements were used to disrupt genes, producing certain phenotypes under various growth conditions that contribute allow a better understanding of the protein's function. This database also contains some expression information and protein localization data.
PDB- This database contains structural information about many proteins. Interactive structure files may be accessed for proteins whose structures have been determined.
Yeast Two-Hybrid Analysis- This website contains information about protein-protein interactions in Saccharomyces cerevisiae. The data was obtained by yeast two-hybrid analysis, a method in which two interacting proteins activate transcription of a reporter gene (See a webpage with more information about this method). Supplementary data has also been made available since the first publication.
DIP- This Database of Interacting Proteins is another source of information about protein interactions, compiled from many different sources. Protein interactions are displayed as networks. The legend for these circuit diagrams that will appear in this webpage is shown below:
: 
Image from: http://dip.doe-mbi.ucla.edu/dip/Search.cgi
PROWL- This database allows the user to search proteins in many different species and then contains links to BLASTP, related proteins, and sequence analysis. The sequence analysis allows the calculation of the molecular weight and isoelectric points of proteins.
Swiss-2DPAGE- This database contains data from two-dimensional (2D) gel electrophoresis for many species, including Saccharomyces cerevisiae. If the spot for a protein of interest was identified on a gel, then the protein is known to be present in the proteome at the particular time and under the particular conditions that the 2D gel was run.
WIT- This database, What is There?, provides a large compilation of information about ORFs, metabolic pathways, enzymes and ortholog clusters.
MIPS: CYGD- This database, the Comprehensive Yeast Genome Database, sponsored by the Munich Information Center for Protein Sequences, provides much information about yeast protein function.
Schwikowski Figures- This page contains links to a large figure displaying protein-protein interactions.
Review of GPA1 Function
In the previous webpages, I have shown that GPA1 is active in the mating signal transduction pathway in Saccharomyces cerevisiae in the following ways:
- GPA1 acts as the alpha subunit of the heterotrimeric G protein involved in the mating cascade and its expression is thus induced in response to mating pheromone treatment.
- This alpha subunit interacts with integral membrane mating pheromone receptors and participates in GTPase activity.
- When stimulated by a mating pheromone, GPA1 releases the beta and gamma subunits of the G protein to initiate the signaling cascade that leads to cellular differentiation, cell cycle arrest and the formation of an oblong cell shape called a "shmoo."
- GPA1 itself appears to play an inhibitory role in the mating response, balancing the stimulatory effects of the beta and gamma subunits of the G protein and allowing cells to adapt to the mating signal and resume the cell cycle if no fusion occurs.
GPA1 Proteomic Data
Protein Function Information:
TRIPLES Deletion Experiment
The image below shows the insertion of an mTn element into GPA1 in the knockout experiment.

Image from: http://ygac.med.yale.edu/triples/execqry.asp?server=oracle
This disruption provided no phenotype information that differed from the wild type. This is likely due to the fact that GPA1 does not play an active role in the conditions tested in the macroarray analysis. A condition in which the mating pheromone response was tested would provide better information about the effect of this disruption on GPA1 function. Though no strong conclusions may be drawn from a negative result, the lack of difference from wild type in these phenotype tests tends to confirm the statement that GPA1 plays a role in the mating pathway and does not play a signaling role in pathways such as cell wall biogenesis, oxidative phosphorylation, etc. that were tested in these experiments (Ross-MacDonald, et al., 1999).
Swiss 2D PAGE Database- This database contained no information for GPA1.
Protein Structure Information:
PDB
No structure information was found at PDB using any of the synonyms GPA1, DAC1, SCG1, CDC70, or YHR005C. Nothing was returned for a search of "G protein alpha subunit". This suggests that the structure of this protein has not been determined. This may be due to the difficulty of isolating large amounts of purified protein due to its association with the plasma membrane. Structures of homologs?
PROWL-
The entry for GPA1 at this website permits sequence analysis. The analysis calculates the molecular mass of GPA1 as 54041.637 Da, and the isoelectric point as 7.7.
Protein-Protein Interactions:
From the information previously obtained about GPA1, it would be expected that GPA1 would interact with the beta and gamma subunits of the heterotrimeric G protein (STE4 and STE18), the pheromone receptor (STE2) and perhaps the kinase FUS3. Various databases that detail protein-protein interaction data obtained by multiple experimental methods can test this prediction.
Two-Hybrid Analysis
No information about protein interactions of GPA1 was given in the initial results from the yeast two hybrid analysis, but in the more recent additions, which occurred after the paper was published, proteins interacting with GPA1 were documented as shown below:
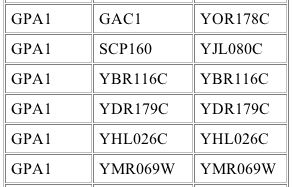
Image from: http://depts.washington.edu/%7Eyeastrc/th_12.htm
The leftmost column indicates that GPA1 was the bait protein. The middle and right columns indicate the prey gene name and the prey ORF respectively that interacted with the bait (Uetz, et al., 2000). The identities of these interacting proteins are shown below:
Gene Name |
Molecular Function |
Biological Process |
Cellular Component |
| GAC1 | protein phosphatase intrinsic regulator activity | Regulates phosphoprotein phosphatase type 1 which in turn regulates glycogen synthase-2 | protein phosphatase type 1 complex |
| SCP160 | RNA binding | Likely controls chromsomes and partitioning of the ER-nuclear membranes during cell division | nuclear envelope-ER network |
| YBR116C | unknown | unknown | unknown |
| YDR179C | deneddylation of Cdc53p | adaptation to pheromone during conjugation with cellular fusion |
Cop9 signalosome complex |
| YHL026C | unknown | unknown | unknown |
| YMR069W | unknown | unknown | cytoplasm, nucleus |
Data from SGD
Interestingly, this experiment gave none of the expected interacting proteins. The interaction of GPA1 with YDR179C was not seen in the simple signaling pathway, but does support the conclusion that GPA1 plays a role in adaptation to the mating signal. The interactions with the other proteins are less easily explained. Perhaps GPA1 regulates these proteins involved in glycogen synthesis and cell division control as it downregulates the mating response. Further experimentation is needed both to verify these interactions and to explain them.
DIP
The graph below shows the interaction map for GPA1 obtained from the DIP database:
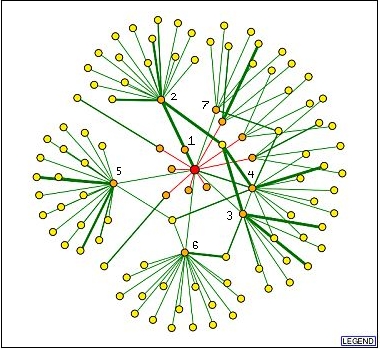
Image from: http://dip.doe-mbi.ucla.edu/dip/DIPgraph.cgi
The identities of the orange nodes (directly interacting with GPA1) which are connected with green lines (verified by one or more methods) are as follows:
1: NAT4/YMR069W- The interaction between GPA1 and this protein was the most strongly verified and was also seen in the yeast two-hybrid experiment. Unfortunately, this corresponds to a hypothetical ORF with unknown biological process and molecular function. It is known to be located in the nucleus and cytoplasm, consistent with the known location of GPA1 in the cytoplasm.
2: STE4- This protein is beta subunit of the heterotrimeric G-protein and is expected to interact with GPA1.
3: FUS3/YBL016W- This protein is involved
in MAP kinase activity to signal cell cycle arrest and to participate in
signal transduction during conjugation with cellular fusion. This interaction
further confirms other research which has suggested that GPA1 interacts with
FUS3 to prevent its action and the
continuation
of the mating response (Blackwell, et. al., 2003).
4: STE11/YLR362W- This protein is involved in kinase activity to cause pseudohyphal growth and signal transduction during conjugation with cellular fusion. Though this protein was not predicted to interact with GPA1 in the simplified signaling pathway, it is reasonable that a protein involved with the mating signal transduction pathway would interact with GPA1.
5: UBI4/YLL039C- This protein is involved in protein degradation tagging activity, protein deubiquitination, protein monoubiquitination, protein polyubiquitination during the response to stress and sporulation. This interaction is unexpected, but may complement information about noticeable changes in GPA1 expression levels during the stress response that was cited in a previous webpage. Perhaps GPA1 plays a yet uncharacterized role in the adaptation to stress as well as the adaptation to the mating response.
6: SST2/YLR452C: This protein is involved in GTPase activator activity and adaptation to pheromone during conjugation with cellular fusion. Like the interaction with YDR179C seen in the yeast two hybrid experiment, this interaction again supports the role of GPA1 in encouraging the adaptation to mating pheromone.
7: SSA2/YLL024C: This protein is involved in SRP-dependent cotranslational membrane targeting, and protein folding. This protein may interact with GPA1 after GPA1 is translated to assist in its targeting to its location near the plasma membrane.
Protein information from SGD (Dolinski, et al., 2003).
Notably, the integral membrane receptors that are known to interact with GPA1 are not seen in these data. This emphasizes that the methods for determining protein interactions are far from perfect, and caution must be used in drawing conclusions from this information.
Schwikowski Diagrams
Interaction diagrams produced by a collaborative effort present the interactions of GPA1 in a slightly more complex diagram. The image shown below is a subset of a large protein interaction diagram (Schwikowski, et al., 2000).
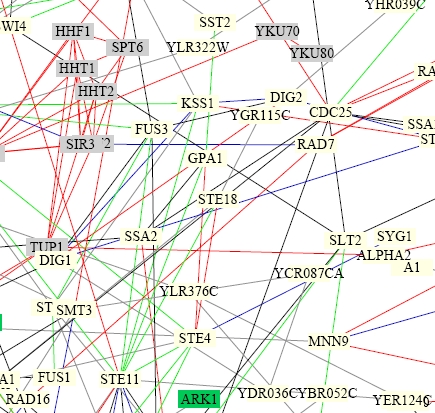
Image from: http://depts.washington.edu/sfields/yplm/data/NB_Figure1color.pdf
Legend-
For lines connecting proteins: red = cellular role and subcellular localization of interacting proteins are identical; blue = localiations are identical; green = cellular roles are identical; black = cellular role and localization are different or unknown
Though it is difficult to tell exactly which proteins are linked to GPA1 (near the center), it is evident that, as shown previously, GPA1 interacts with the G protein beta subunit STE4, the kinase STE11, and the mating pheromone adaptation protein SST2. This figure also makes clear the reason that the gamma subunit of the G protein, STE18, had not been seen on the other interaction diagrams. The red lines in this figure show that GPA1 only directly interacts with STE4, the beta subunit, which in turn interacts with STE18.
Experiments to Test GPA1 Function
As cited on an earlier page in which the effects of polymorphisms in the GPA1 gene were investigated, many experiments have already been done to test the basic function of GPA1. Further experiments should work to determine its precise role in promoting the mating response, to test its role in the adaptation to the mating pheromone signal, and to investigate any other possible roles in the life of yeast.
- An experiment to characterize the effect of the mTn insertion in GPA1 from the TRIPLES experiment would help to confirm the role of GPA1 in the mating pathway. Using methods similar to those that the original authors used, phenotype macroarrays would be prepared with cells in the presence and absence of alpha or "a" mating pheromone (Ross-MacDonald, et al., 1999). Cells which have transposon insertions in genes involved in the mating pathway should show altered growth from wild type under pheromone exposure. Wild type cells would be expected to show reduced growth when exposed to mating factor as they undergo cell cycle arrest and form shmoos. Some growth will be seen however, as proteins like GPA1 permit adaptation to the mating signal. Cells in which GPA1 is disrupted by the mTn insertion should thus show decreased growth relative to wildtype due to the lack of adaptation to pheromone.
- During several events in the life of yeast, GPA1 is known to be located near the plasma membrane to interact with pheromone receptors. However, an experiment using GFP labeling of GPA1 would allow researchers to monitor the localization of this protein through the yeast life cycle. This would allow the cellular component of GPA1 to be identified during both its initial signaling role and during its role in adaptation to the signal.
- The exact mechanism of GPA1 function could be better explained if the structure, or at least the specific binding sites of the protein were understood. This could be further explored by identifying potential binding domains and active sites using the protein sequence and structure predictions. Using this information, inhibitors specific to each class of active sites could be included one at a time in the yeast two hybrid interaction experiment. If an inhibitor blocked an interaction site, an interaction previously seen between GPA1 and a second protein would no longer occur. This would give an indication of what kind of binding domain interacted with each protein (STE4, FUS3, etc.). Each inhibitor could also be incubated with the GPA1 protein in a microwell array containing radioactive GTP, similar to the method used in Snyder's lab, to determine which inhibitor blocked the GTPase activity of the protein (Zhu, et al., 2000).
Review of YHR003C Function Prediction
In the previous webpages, the analysis of DNA sequence, conserved domains, homologs, and expression patterns of YHR003C gave the following information:
- YHR003C appears to be a globular protein and shows much sequence homology to dinucleotide-utilizing enzymes that are involved in molybdopterin and thiamine biosynthesis.
- YHR003C contains conserved ThiF domains that, in other proteins allow function in mRNA binding or mitochondrial protein transport.
- During several conditions, YHR003C shares expression patterns with known genes whose proteins are involved in ribosome biogenesis, metabolism, biosynthesis, and RNA processing.
- YHR003C is repressed when the yeast cell enters stationary phase, as expected for a protein involved in biosynthesis.
From this information, I previously predicted that YHR003C was a globular protein which functions as an enzyme involved in ribosome biogenesis and metabolism. It may have catalytic activity involving nucleotide binding.
YHR003C Proteomic Data
Protein Function Information:
TRIPLES Deletion Experiment
This table shows that there were multiple transposon insertions into the YHR003C gene at different points. In one case, it appears that the insertion affected a stop codon so that the protein had a much extended length. "In-frame" indicates that the LacZ protein is in the correct reading frame when inserted into the gene.
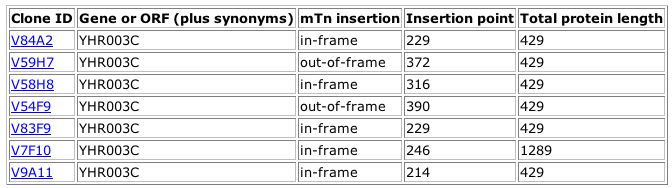
Image from: http://ygac.med.yale.edu/triples/execqry.asp?server=oracle
For all but the last clone, the yeast cells showed wild type growth patterns on the phenotype macroarray analysis. This indicates that the disruptions did not cause changes that caused the cells to respond differently under many conditions. For clone V9A11, the HHIG growth condition produced a phenotype different from wildtype as shown in the table below:
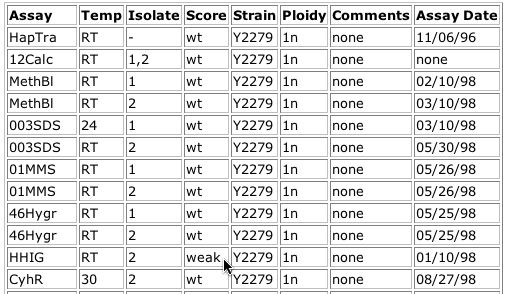
Image from: http://ygac.med.yale.edu/triples/get_clone_info.asp?cloneid=V9A11
The HHIG assay, according to the key, was a test for hyperhaploid invasive growth mutants. This phenotype was detected by growth of yeast on normal YPD medium at 24 degrees celsius for 6 days. The classification "weak" indicates that the growth pattern was only slightly different from wild type. At a basic level, the observance of a differing phenotype under a certain condition when this hypothetical ORF is disrupted confirms that this ORF actually is a yeast gene (Ross-MacDonald, et al., 1999).
Other genes that produced a phenotype under this condition when disrupted were:
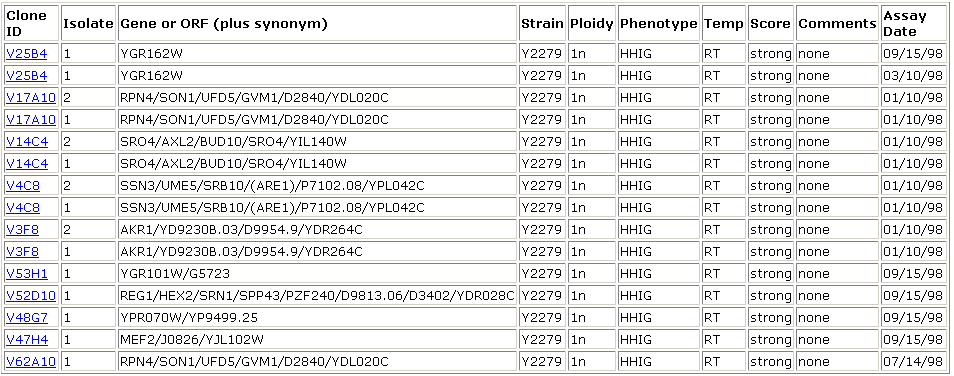
Image from: http://ygac.med.yale.edu/triples/execqry.asp?subsql=yes&server=oracle
According to the Saccharomyces Genome Database, several of these genes (YGR162W and YPR070W) are involved in transcription and translation. Many others are involved in processes of metabolism and biosynthesis. YGR101W, for example, is involved in mitochondrion organization and biosynthesis. REG1 is involved in cell growth and RPN4 is involved in protein catabolism. Since YHR003C shows a similar (albeit weaker) phenotype under the same conditions as these proteins, it may play a similar role. Both this information and the fact that a protein affecting invasive growth is likely involved in biogenesis support previous hypotheses about YHR003C. Several of these proteins are located in the mitochondrion, raising the question again of whether YHR003C might be localized in or near the mitochondrion.
Swiss 2D PAGE Database- YHR003C was not identified on the 2D gel for yeast.
What Is There?
The WIT database cites the function of YHR003C as molibdopterin and thiamine biosynthesis, based on its conserved domain information.
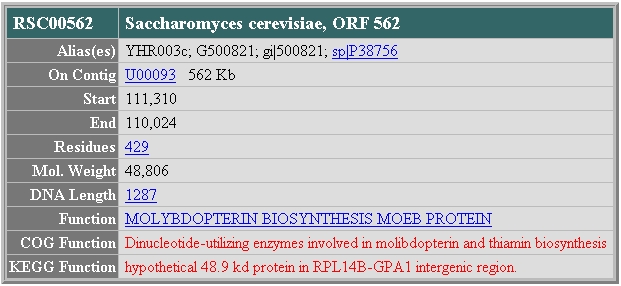
Image from: http://wit.mcs.anl.gov/WIT2/CGI/prot.cgi?prot=RSC00562&user=
MIPS
The comprehensive yeast genome database also provides information in which the functional category of YHR003C is listed as "metabolism" and biosynthesis. This information is also based on similarities to known proteins.

Image from: http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YHR003c
Protein Structure Information:
PDB
No structure files were found in the Protein Database with a search for YHR003C. Also, neither the Saccharomyces cerevisiae homolog nor the Schizosaccharomyces homologous protein had a structure file.
Protein-Protein Interactions:
Two Hybrid Analysis
No information was given in the initial or updated results.
DIP
The graph below shows the interaction map for GPA1 obtained from the DIP database:
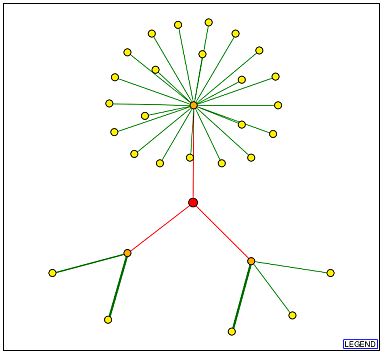
Image from:http://dip.doe-mbi.ucla.edu/dip/DIPgraph.cgi?PK=4417&D=2
It is interesting to note from the above diagram that there are only three connections to the YHR003C node (red) while there were more than ten to the GPA1 node shown earlier. Also, each of these connecting edges is red, indicating, as shown in the legend, that these interactions are unverified results of high-throughput screens. These data may indicate that YHR003C is not a central player in a protein network, or it may simply indicate that less work has been done with this protein. Some useful information may be gathered from the identities of the three proteins with which YHR003C is thought to directly interact, however. These are represented by the orange nodes on the graph above and have the following identities (counterclockwise from top node):
| Protein Name | Molecular Function | Biological Process | Cellular Component |
| JSN1/YJR091C | mRNA binding | deadenylation-dependent mRNA catabolism | unknown |
| DSE1/YER124C | unknown | cell wall organization and biogenesis | bud neck |
| TOM40/YMR203W | protein transporter activity | mitochondrial translocation | mitochondrion outer membrane translocase complex |
Data from: SGD
The large cluster of proteins interacting with JSN1 have several different functions. Several are hypothetical membrane proteins, others are protein kinases (such as MKK2). Most of these proteins are located either at the membrane or in the cytoplasm. This suggests that the cellular component of JSN1 is the cytoplasm. Both proteins associated with DSE1 are involved in Rho protein signal transduction for the establishment of cell polarity during budding. All three proteins that interact with TOM40 are involved in mitochondrial translocase complexes on either the inner or the outer mitochondrial membrane.
These interacting proteins have quite diverse functions and even locations within the yeast cell, so it is difficult to understand how YHR003C might interact with all three. However, the hypothesis that YHR003C is an enzyme involved in metabolism and biogenesis is supported by its interaction with proteins involved in cell wall biogenesis and mRNA catabolism. If YHR003C does indeed interact with all of these three proteins, it must be located in the cytoplasm and travel from locations near the plasma membrane to locations near the mitochondria.
The MIPS Protein-Protein Interaction Database lists the same three interacting proteins.

Image from: http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YHR003c
Schwikowski Diagrams
YHR003C was not included on any of the large protein interaction diagrams produced by Schwikowski, et al.
Revised Prediction of YHR003C Function
As when the expression data and gene data were analyzed, the proteomic data suggest that YHR003C is involved in biosynthesis. However, though this basic functional category is better confirmed by the proteomic data, the precise function is less clear after analyzing this data than it was when considering the gene expression data. Certain combinations of protein interactions have made the protein localization uncertain and the precise biosynthetic process is also unclear.
Experiments to Test Prediction of YHR003C Function
Though predictions from high-throughput database information are important, definitive conclusions about the function of the non-annotated protein can only come from careful laboratory experiments.
- One of the most uncertain aspects of YHR003C is its localization. Some data suggest that it is located in the cytoplasm while other data indicate that it is located in some portion of the mitochondria or near the cell wall. The localization data given in the TRIPLES epitope tag experiment gave no data different from background, so a different approach must be tried. One method that would allow the protein localization to be visualized in the yeast cell would be to tag the YHR003C gene with Green Flourescent Protein (GFP). It would be expected that this labeling would produce green fluorescence either in the cytoplasm, confined to the cell wall, or within the mitochondria.
- In the case of GPA1, much information about its functional role was obtained by the analysis of mutant phenotypes. More information could be obtained about YHR003C by studying similar polymorphisms. Many cells could be screened for polymorphisms in the YHR003c gene by sequencing this locus. The phenotypic manifestations of these polymorphisms could then be analyzed. If YHR003C is indeed involved in biosynthesis, a process in which vital larger molecules are made, then a defect in this gene should result in decreased cell growth when cells are cultured with ample space and growth medium.
- Another remaining uncertainty about the function of YHR003C is determining the precise process of metabolism and biosynthesis in which it participates. The data from these online databases present various choices: ribosome biogenesis, cell wall biogenesis, vitamin and cofactor biosynthesis, or biosynthesis in the mitochondrion. If YHR003C is disrupted, as in the transposon interuption or in another mutant phenotype determined in the type of screen described above, one or more of these pathways should be deficient. To determine whether YHR003C played a role in vitamin and cofactor biosynthesis, mutant and wild type yeast metabolomes could be compared at several different time points during the cell cycle. The mutation in YHR003C would be expected to cause the concentrations of certain nonprotein compounds as determined by gas chromatography and mass spectrometry to be altered significantly in the mutant as compared to the wild type cells (Fiehn, et al., 2000).
References
Blackwell E. et. al. 2003. Effect of the pheromone-responsive G(alpha) and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase. Mol Cell Biol. 23: 1135-50. <http://mcb.asm.org/cgi/content/full/23/4/1135?view=full&pmid=12556475> Accessed 2003 4 Oct.
Dolinski, K. et al. (2003). Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed 2003 8 Nov.
Fiehn, O. et al. 2000. Metabolic profiling for plant functional genomics. Nature Biotechnology 18: 1157-1161.
Ross-Macdonald P. et al. (1999). Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 402: 413-8.
Schwikowski, B. et al. (2000). A Network of Protein-Protein Interactions in Yeast. Nature Biotechnology. 18: 1257-1261.
Uetz P. et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403: 623-7.
Zhu, H. et al. (2001) Protein Arrays and microarrays. Current Opinion in Chemical Biology 5: 40-45.
Questions or Comments? Please e-mail Rachel Patton McCord
Rachel Patton McCord's Genomics Homepage