*This web page was produced as part of an undergraduate course at Davidson College.*
My Favorite Yeast Expression
This is a page dedicated to further analysis of yeast genes individually selected by our genomics class through the use of DNA microarrays. (DNA Microarray Methodology - Flash Animation, 2003, <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/method/chip/chip.html>)
Annotated Gene- FUR4
The gene FUR4 is a member of a family of genes that is located on chromosome 2 in Saccharomyces cerevisae that serves to produce uracil permease. In order to better orient yourself with FUR4, its functioning, and uracil permease see my Favorite Yeast Gene page. Here, I will offer a brief description in terms of gene ontology:
Biological Process- FUR4 produces uracil permease, a specific co-transporter of uracil across the cell membrane in yeast cells. It is a membrane-spanning protein and bears a similar structure to many others of its kind in existence.
Molecular Function- Uracil permease is a symporter enzyme within the plasma membrane. It generates an electrochemical gradient across the membrane that is selectively permeable to uracil and binds it before quickly releasing it within the membrane. This gradient is generated by co-transport of protons.
Cellular Component- FUR4's protein product, uracil permease, is integral to the plasma membrane as mentioned before. This is obviously integral to its function and very similar to many other types of membrane transport proteins.
The source of the following microarray results should first be noted. All of the following images were obtained from the Expression Connection (Expression Connection, 2003, <http://db.yeastgenome.org/cgi-bin/SGD/expression/expressionConnection.pl>). In examining the different microarrays that were available, I attempted to select for those that suggested something of particular interest with regards to FUR4 expression.
![]()
Figure 1- This is the color scale that will be used to gauge the repression or induction of each gene in the microarray images that will follow.
Similarly Expressed Genes: Expression during sporulation Chu S, et al.
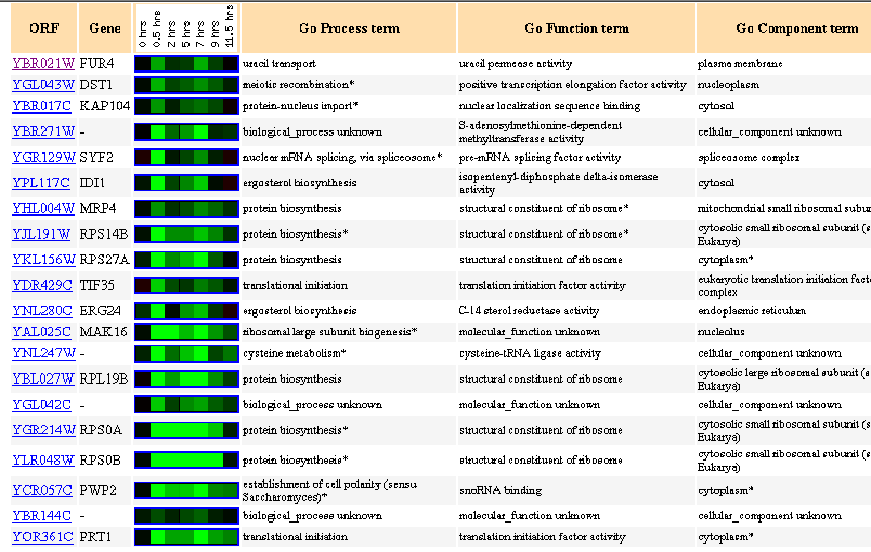
Figure 2- This image depicts the results of a microarray of expression of FUR4 during sporulation and similar genes with a p score of greater than .8.
Sporulation, for yeast cells, is quite simply the formation of spores. FUR4 is repressed at .5 hours in spore production and then again at 7 hours in spore production. While it is unclear as to why FUR4 would be repressed during spore production, what is significant about this figure is the similarity that FUR4 has with other genes. Notice that almost every single gene on the list has something to do with RNA functioning in transcription or translation. This makes perfect sense as uracil permease is involved in the import and export of uracil across the plasma membrane. Uracil, according to basic genetics, is a pyrmidine building block of RNA and would be required in its construction. It would make sense that these genes would be associated in some way, not just in sporulation but in most cellular processes. We also see levels of similar repression at similar times with genes such as IDI1 and ERG24 which are involved in ergosterol biosynthesis, this could be due to uracil's involvement also as a source of cellular energy.
Similarly Expressed Genes: Expression during the diauxic shift DeRisi JL, et al.
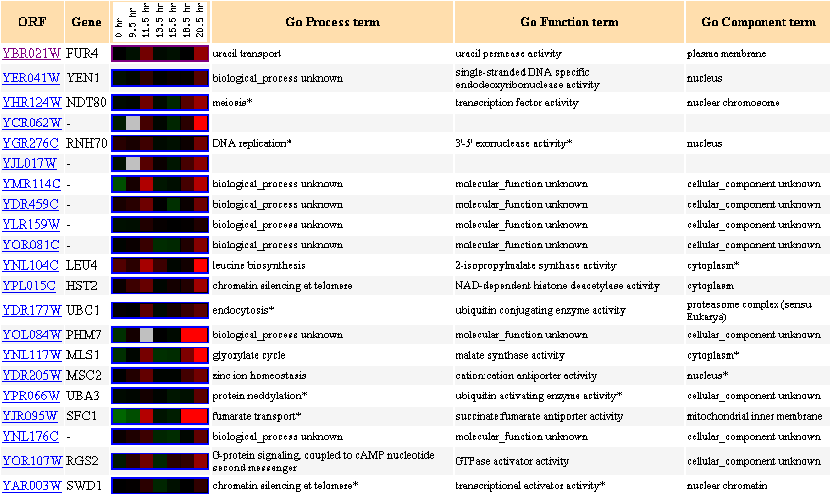
Figure 3- This image depicts the results of a microarray of expression of FUR4 during a diauxic shift and similar genes with a p score of greater than .8.
A diauxic shift in a yeast cell is a switch in respiration type from anaerobic to aerobic respiration. In this experiment a slight induction of FUR4 can be seen at the 11.5 hour mark and then again more strongly at the 20.5 hour mark. Notice again with genes YEN1, NDT80, and RNH70 that the FUR4 gene shares a similar induction profile with other DNA/RNA processing genes. The NDT80 gene involved in meiosis especially caught my eye in its similarity. We also see a similar induction profile in comparison with genes that could have to do with uracil's use as an energy source such as LEU4 or MLS1. Again, what effects a change in respiration type would have on the import or export of uracil is unknown to me but what is indeed expected and observed is the preservation of the shared profile between the FUR4 and the DNA/RNA processing genes.
Similarly Expressed Genes: Expression in response to DNA-damaging agents Gasch AP, et al.

Figure 4- This image depicts the results of a microarray of expression of FUR4 during exposure to different types of DNA-damaging agents and similar genes with a p score of greater than .8.
It is interesting that in this case there were no genes with a p score of greater than .8 to which FUR4 could be compared. However, the response of FUR4 to these different conditions is intriguing. We observe a general pattern of repression in this microarray in the various different experimental conditions but what is interesting is its behavior during gamma irradiation. At the 5 minute mark, a very strong repression can be observed which is followed by a slight induction through the rest of the time period. Again, a similar pattern of behavior can be observed during mock gamma irradiation of wild type FUR4. This time it starts with a slight repression and then gives a very strong inductive signal near the end of the irradiation period at the 60 minute mark. During irradiation, which is basically random severing of strands of DNA, FUR4 is repressed initially due to perhaps a slight halt in DNA/RNA activity due to the damage that it suddenly incurs. This is maybe followed by a compensatory period in which the FUR4 protein attempts to import more uracil for DNA/RNA repairs and thus an induction occurs later on.
Similarly Expressed Genes: Expression in response to alpha-factor (over time) Roberts CJ, et al.
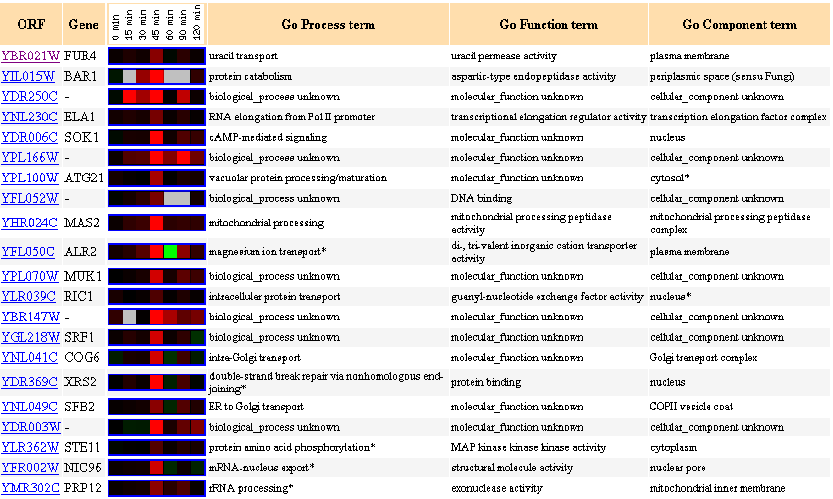
Figure 5- This image depicts the results of a microarray of expression of FUR4 during exposure to alpha-factor and similar genes with a p score of greater than .8.
Exposure to this growth factor shows an induction of FUR4 at around the 45 minute mark. It is not surprising for increased uracil transport activity to occur just before cell growth as more DNA/RNA processing as well as energy usage would be happening just after that initial halt to cell activity that the alpha factor produces. Striking is the similarity between FUR4 and ELA1, ATG21, NIC96, and PRP12- again all genes associated with mRNA or protein processing.
Similarly Expressed Genes: Expression in response to histone depletion Wyrick JJ, et al.
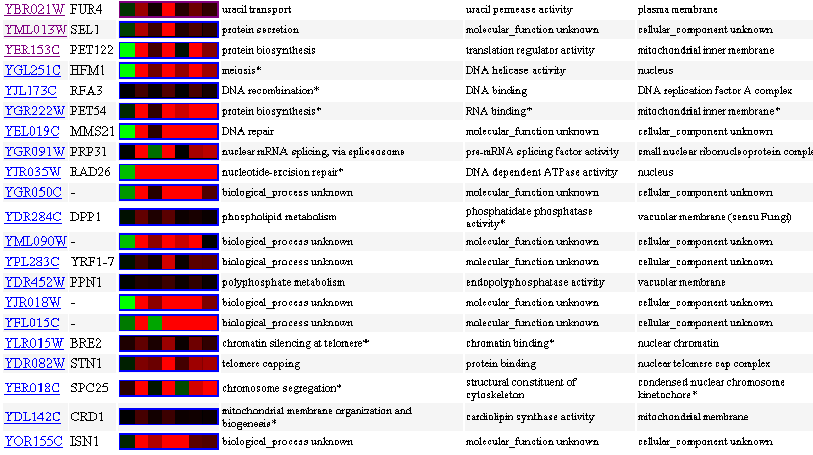
Figure 6- This image depicts the results of a microarray of expression of FUR4 during histone depletion and similar genes with a p score of greater than .8.
In response to histone depletion, building blocks of DNA structure, the FUR4 gene expresses a general trend of induction with its greatest induction at the 1.5 hour mark. Notice also the striking similarity between FUR4 and SEL1, it is known that SEL1 is involved in protein secretion of some type but beyond that not much is known. Due to their similarity, I would venture to guess that SEL1 perhaps secretes a protein that works to accomplish either a similar function to FUR4 or some sort of DNA/RNA maintenance due to the clustered induction and repression of associated genes. It is true that it is also possible that the two are simply regulated by a similar promoter; however, because SEL1 is involved in protein secretion as is FUR4 in a way- I would feel more comfortable making a functional conclusion. Again we see patterns of similar induction among genes related to DNA and RNA maintenance as well as transcription and translation.
Conclusions-
What can be observed here is a simple application of various microarray results to further explore and define the role of a known gene and its protein in cellular function. The FUR4 gene, whose simple yet crucial function is mostly understood, functions within the cellular system much as can be expected it seems. Uracil's role as an RNA building block and involvement as an energy source for cellular processes would logically associate it with genes involved in the regulation of RNA/DNA, transcription, translation, and even in the biosynthesis of certain proteins or sugars. It would therefore make sense also that FUR4, a gene involved in transporting uracil into and out of the cell, reacts along with these genes. This is indeed what we observe over several experiments. Microarrays also allow one to make guesses regarding the function and location of unknown genes with similar expression profiles (assuming the reason they have a similar expression profile is not because they share a common or very similar promoter) such as with the SEL1 protein during histone depletion. The practical applications of these results for genomic research are incredible and it will be interesting to see how far it will carry the scientific community.
Unannotated Gene- YBL010C
In my last web venture, I attempted to characterize and propose some sort of function for the ORF YBL010C as it is located near FUR4 and nothing is known of it save for its sequence. I could only draw two vague conclusions due to motifs found in its sequence: it was not a membrane protein and it was involved in DNA binding. It is my hope that this microarray data will serve to further characterize this vague sequence.
![]()
Figure 7- This is the color scale that will be used to gauge the repression or induction of each gene in the microarray images that will follow.
| Similarly Expressed Genes: Expression during sporulation Chu S, et al. |
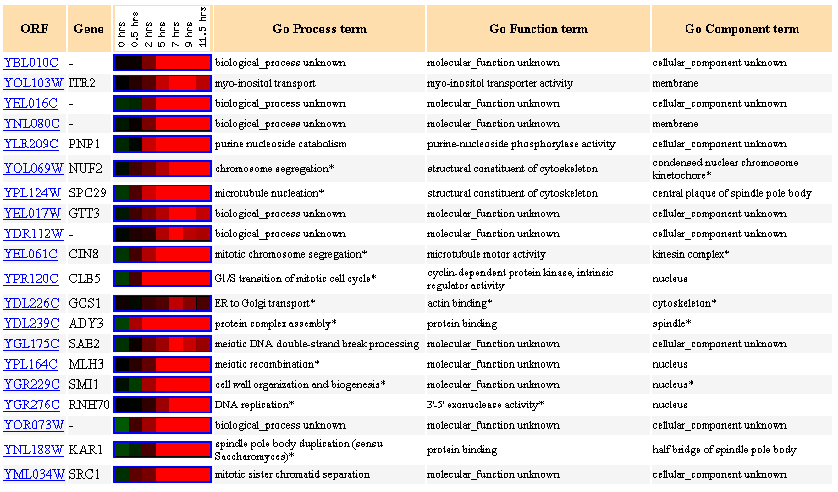
Figure 8- This image depicts the results of a microarray of expression of YBL010C during sporulation and similar genes with a p score of greater than .8.
During sporulation YBL010C experiences a rather drastic induction starting at the 5 hour point. I cannot say for sure why this is but I can propose that since it is involved in DNA binding that the production of spores necessitated increased activity of this ORF's product to produce said spores. I conclude 2 things from this image with regards to the functioning of this ORF. One is that there are proteins present in these results whose similarity of expression to FUR4 I would attribute to a similar promoter such at ITR2. ITR2 is involved in myo-inositol transport and it is located in the membrane, it is known that FUR4 is not in the membrane due to its hydropathy score and thus it is unlikely they are associated in any other way than by promoter. The second thing that I can conclude is that my prediction that the YBL010C protein is involved in DNA binding was not invalidated. NUF2, CIN8, and RNH70 are all genes associated with some sort of structural DNA activity and they each exhibited a strong induction towards the end of the time period. Other genes involved in other types of structural protein binding (cytoskeletal) such as SPC29 also shared similar expression profiles, this is intriguing and might suggest a functional link.
| Similarly Expressed Genes: Expression during the diauxic shift DeRisi JL, et al. |
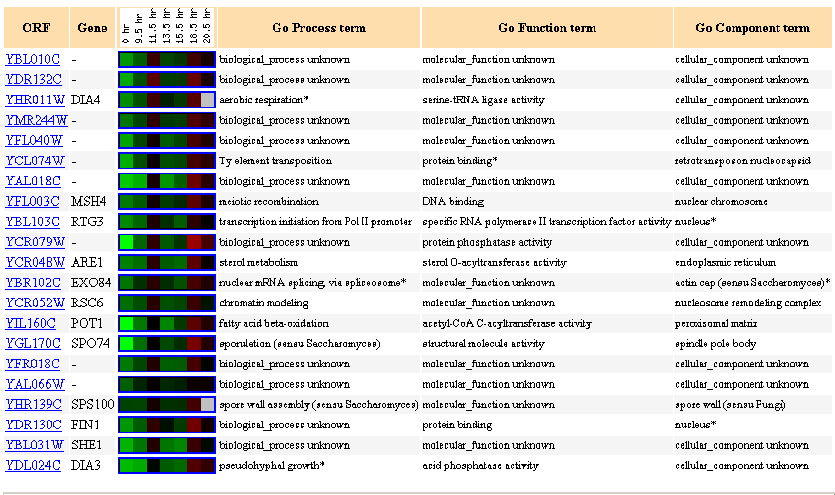
Figure 9- This image depicts the results of a microarray of expression of YBL010C during a diauxic shift and similar genes with a p score of greater than .8.
During a diauxic shift YBL010C is repressed initially but then becomes induced slightly at the 18.5 hour mark. There were many genes in these results with similar expression profiles but unknown functions. What was of particular note to me was the gene MSH4 that is actually a known DNA binding gene. Here we have a known DNA binding gene with a similar expression profile to YBL010C, more evidence of the DNA binding hypothesis.
Similarly Expressed Genes: Expression in response to histone depletion Wyrick JJ, et al.

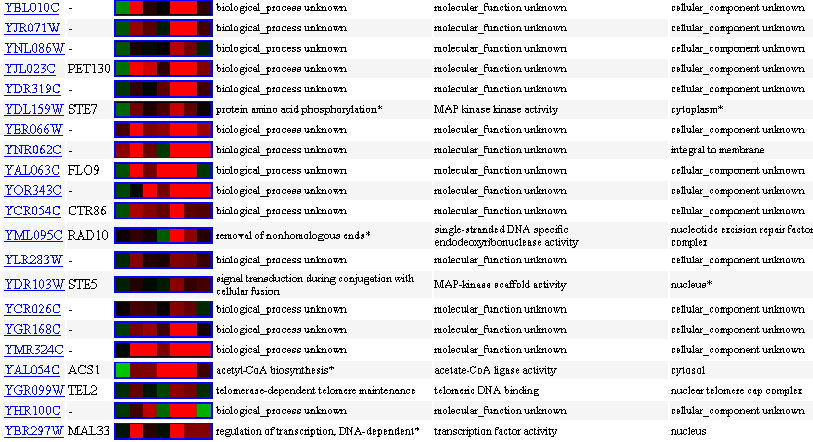
Figure 9- This image depicts the results of a microarray of expression of YBL010C during histone depletion and similar genes with a p score of greater than .8.
The unknown ORF experienced an initial repression and then an induction with strong signals at .5, 2, and 4 hours. There were surprisingly not very many genes about which there are known functions with similar expression profiles. Notice that all but one of the known genes are not located in the cell membrane.
Similarly Expressed Genes: Expression in response to alpha-factor (various concentrations)
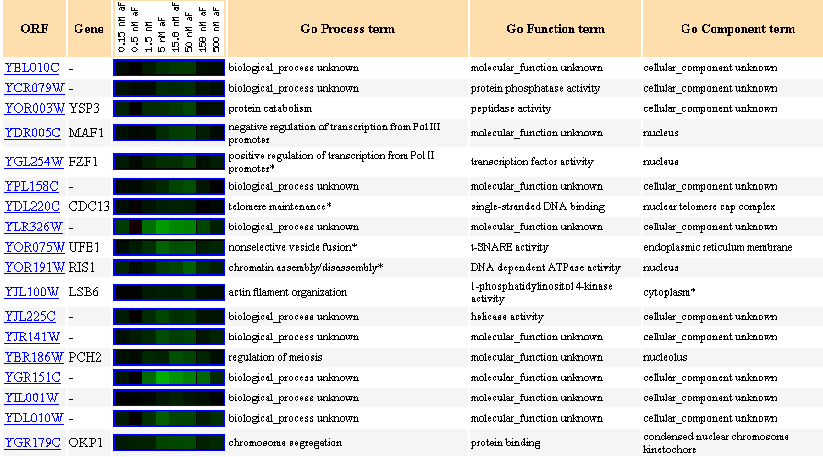
Figure 9- This image depicts the results of a microarray of expression of YBL010C during exposure to alpha factor and similar genes with a p score of greater than .8.
The ORF shows a slight repression in response to exposure to alpha-factor at these different concentrations. The first thing I noticed were the locations of each of the genes with similar expression profiles- all but one are in or in close proximity to the nucleus. Notice the expression similarity of CDC13, OKP1, and RIS1 among others, which all share structural DNA functioning.
Conclusions-
If anything, the results of the different microarray experiments served to give further credence to my original hypothesis. The results neither conclusively proved or disproved what I thought before. The experiments showed that genes involved with DNA structure or binding had very similar expression patterns to the ORF in a number of different variable conditions. One thing that was confirmed without a doubt is that this ORF was associated with the inner area of the cell, perhaps even the nucleus. In a number of the tests a majority, with a few exceptions, of associated genes were not associated with the membrane and many of those with specifically the nucleus or nucleolus. It is not unreasonable then to adhere to my original hypothesis. Based on the results; however, I feel that I might be able to make a somewhat premature but not out of the question proposal. The behavior of YBL010C during sporulation is extremely striking and it displays a strong signal of induction as noted before. Keeping this in mind and its possible association with DNA structural functionality as well as protein binding, I believe that YBL010C might be related to the formation of spores but more specifically to the binding of the DNA during the formation of spores. It may have further implications for any type of yeast cell growth but this will not be known without further experimentation.
References
DNA Microarray Methodology - Flash Animation, 2003, <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/method/chip/chip.html>
Expression Connection, 2003, <http://db.yeastgenome.org/cgi-bin/SGD/expression/expressionConnection.pl>
Return to Matt's Genomics Page
Return to Dr. Campbell's Genomics Homepage
Return to the Davidson College Biology Homepage
Email Matt at matalbert@davidson.edu