
This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Proteins: SKI6 and YGR201C?
Proteomics is a field that promises to bridge the gap between genome sequence and cellular behavior. It aims to study the dynamic protein products of the genome and their interactions, rather than focusing on the simple static DNA blueprint of a cell. Its rapid emergence in biotechnology is being driven by the development, integration, and automation of large-scale analytical tools... Apart from its use in providing fundamental insights into the molecular basis of a cell’s state, the ability to reveal how protein cascades change as a result of specific disorders and drug treatment promises to galvanize the identification and validation of drug targets. (Dove, 1999)
Introduction
This page is designed to explore the functions of my favorite proteins. Many proteomic analysis tools are available online as databases and PDF files. On this page I use these tools to interpret the protein functions and structures encoded by SKI6 and YGR201C.
| Links to Useful Tools |
| MIPS: CYGD |
| PDB |
| PUMA2 |
| Swiss-2DPAGE |
| TRIPLES |
 |
| What is a proteome? This boxed definition was taken from Alan Dove's review of proteomics published in 1999. |
Analysis of SKI6: Structure, Functions, and Interactions
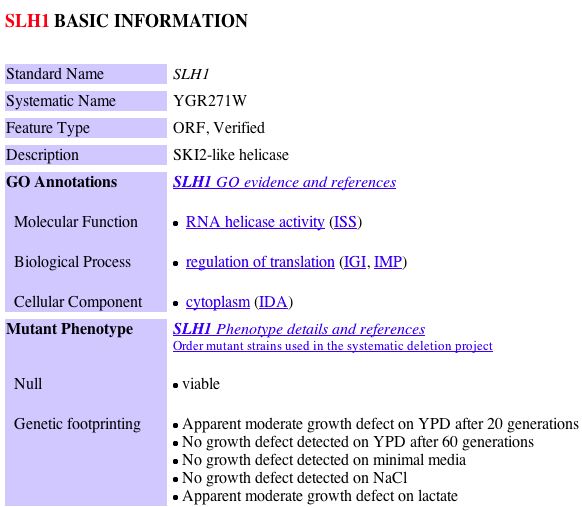
TRIPLES - a database of TRansposon-Insertion Phenotypes, Localization, and Expression in Saccharomyces
| TRIPLES data indicates the results of deleting a protein under various experimental conditions. |
| For each of the six clones tested it was apparent that the mTn insertion for ski6 during sporulation and vegetative states each turned the gel faint/light blue, indicating no active induction under these conditions. |
PROWL- this database is useful in analyzing protein sequence and indicating the existence of relationships among proteins.
| PROWL database results for ski6 showing two related proteins. |
The molecular weight for Ski6 is 27543.464 Da and the isoelectric point is 7.9. The PROWL database also confirmed the identity and function of my known protein. |
DIP- This Database of Interacting Proteins provides links to most known interactions for a given protein.
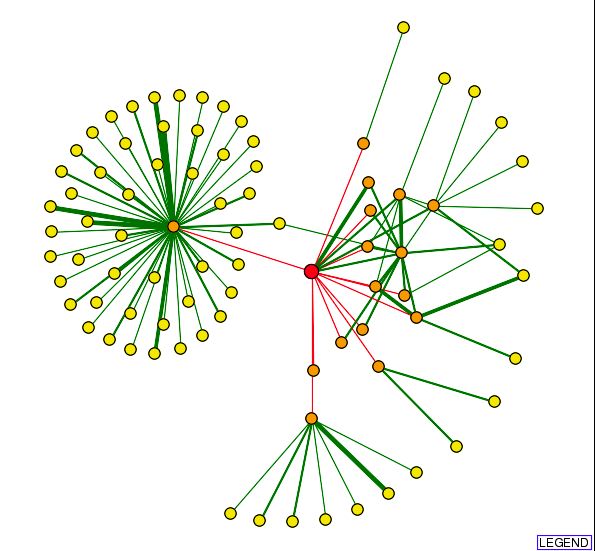
| In the center of the circle to the left is Srp1, a gene involved in nucleocytoplasmic transfer and protein activity. The green lines branching to the right connect to other RRP proteins involved in 3'->5' exoribonuclease processing activity. |
Swiss 2D Page for Ski6 This search failed to yield any certain temporal or location results. The 2-D maps provided only predictions of where this protein might be found. However, we know ski6 functions in RNA processing and the creation of the exosome complex from this and previous data.
I could not find any useful information at the PUMA2 site for Ski6.
PDB found neither information nor structure matches for SKI6 (aka RRP41, ECM20, G7587, or YGR195W).
YRC Two-Hybrid Analysis No protein interactions or activations found for ski6 in this data set.
SKI6 Connections: The PDF files from Benno Schwikowski & Peter Uetz show protein-protein interactions. These connection maps are available in the public domain provided useful information about ski6.
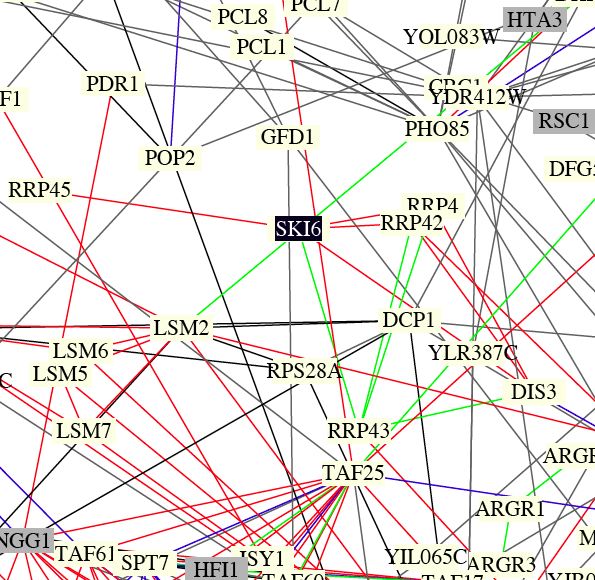 |
| Benno Figure 1. Green lines indicate cellular roles are identical but localizations differ. Red lines mean cellular role and subcellular localization of interacting proteins are identical. Gray boxes mean cromatin structure. (Key found in Campbell page 181) Over 2000 protein interactions are represented in the full figure. Circuit diagrams, though not perfect, provide a way to visualize connections. |
| This figure shows that ski6 has localization identical to the other RRP proteins, confirming the connections from the DIP graphs that were discussed previously. Also, about 10 primary connections (like in DIP) are shown in this figure, providing evidence that the functional connections are significant replicable. |
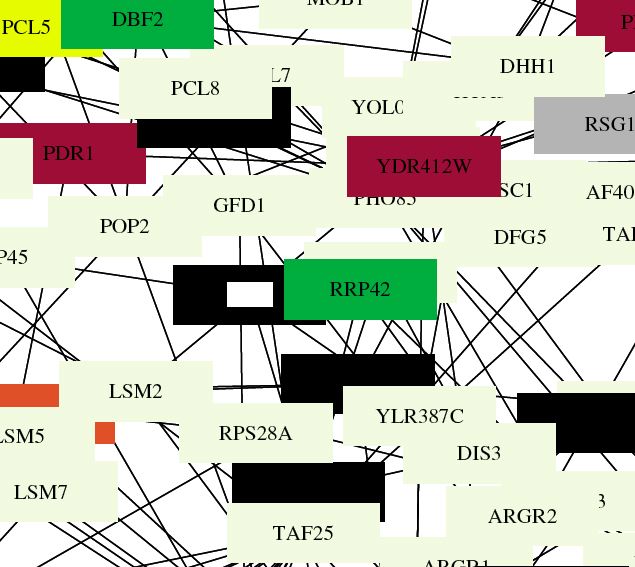 |
 |
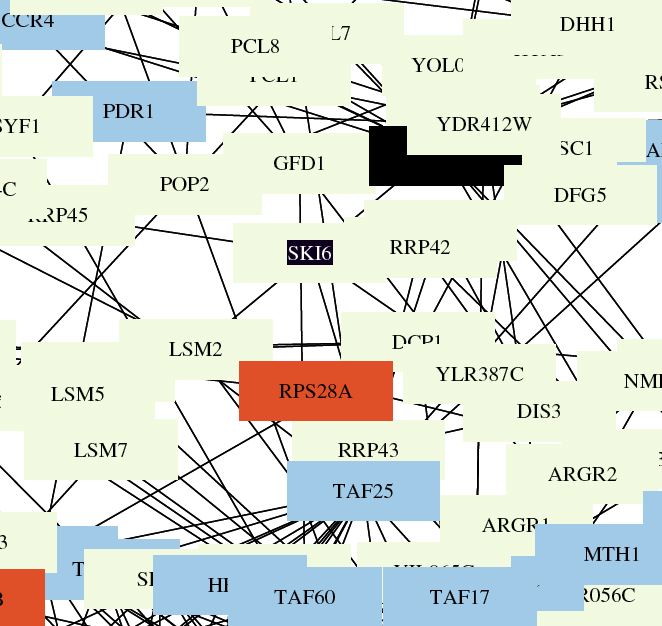
| This figure illustrates the functions of interacting proteins. Ski6 is indicated by the black and white empty box in the center of the map. Ski6 and many of the proteins it interacts with function with RNA-turnover. This is logical since my gene is known to play a role in formation of ribosomes and exosomes related to RNA processing. The format and spacing of this map make it difficult to determine what connections actually exist. |
 |
 |
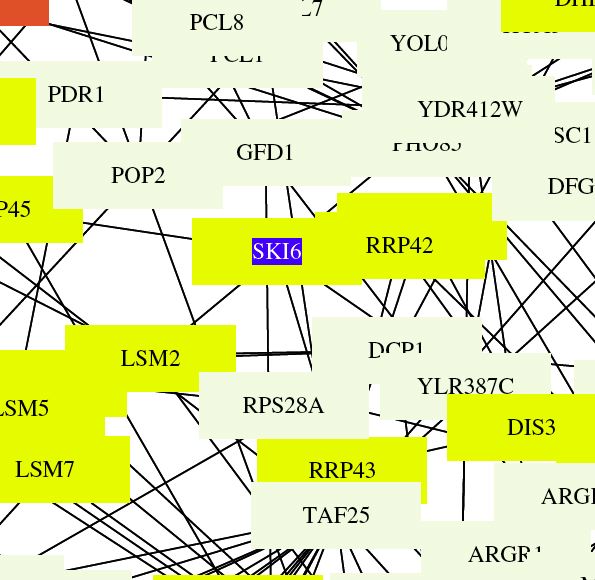
| This figure shows that Ski6 has a definitive roll in RNA processing and modification. Ski6 and all of the surrounding boxes are yellow. Previous experiments listed on the SGD site confirm this data. |
 |
 |
Information from MIPS
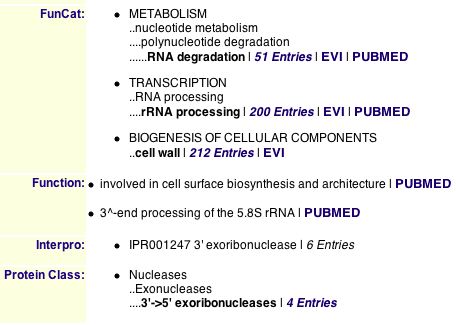

| The roles of Ski6 are listed on these screen shots. By performing a search, I found other proteins that function in the degradation of RNA transcripts. RRP4 and MSU1 are consistently near SKI6 on interaction trees. |
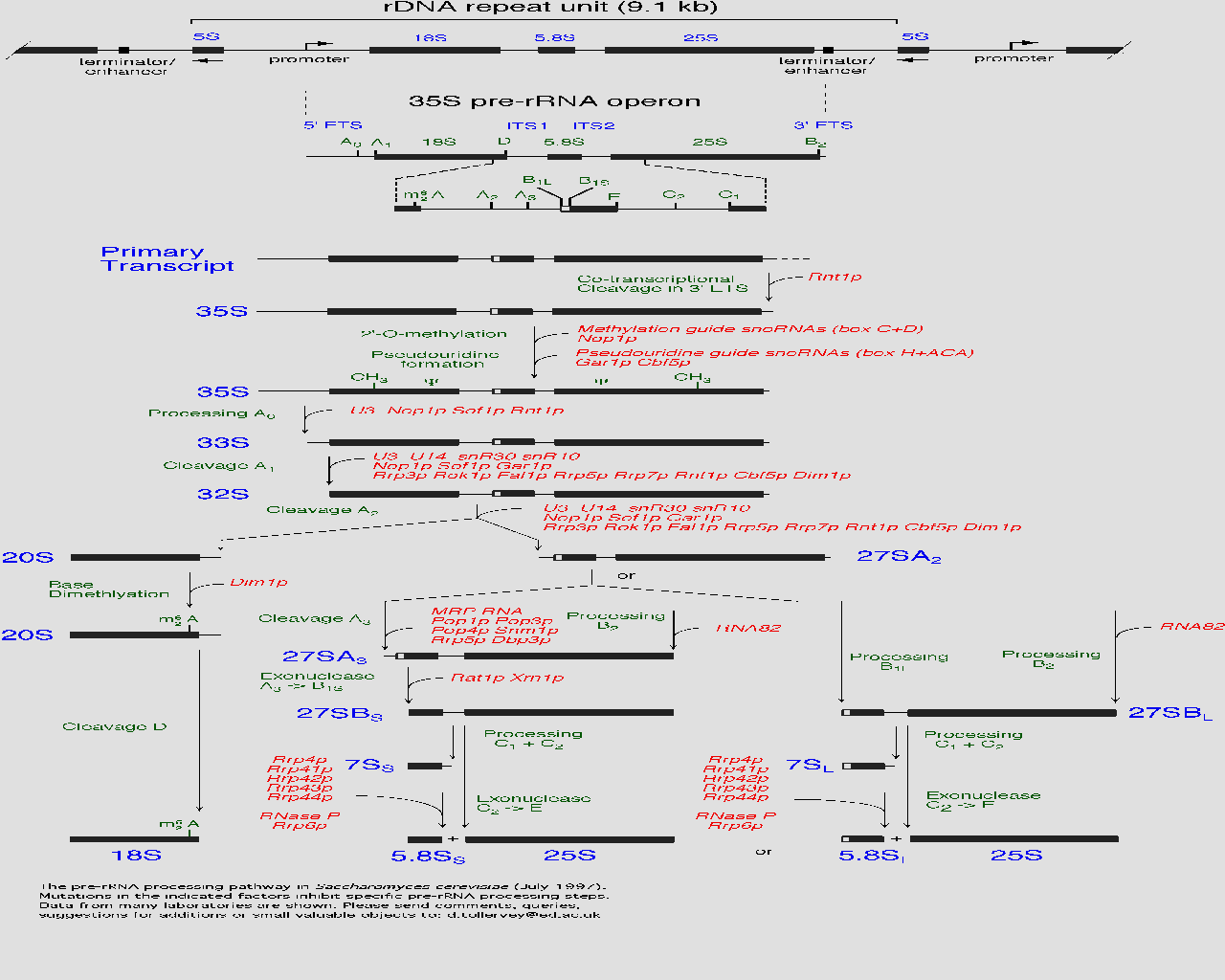
| This diagram found at the MIPS site shows the pre-RNA processing pathway for yeast. Near the bottom of the chart Ski6 appears (as RRP41) with related proteins. My protein is critical in the formation and function of the 5.8 ribosomal RNA subunit. |
I found no data that contradicted the established function of Ski6 during this investigation. I believe my protein plays an important roll in 3'-5' RNA transcript degradation and also helps to form a ribosomal subunit in yeast. It was interesting to discover the other RRP proteins that interact closely with SKI6.
Analysis of YGR201C: Structure, Functions, and Interactions
None of the PDF maps displaying protein-protein interactions included YGR201C. This probably means that my unknown has not been tested or plays a minor role in the processes shown in interaction files.
A search of Swiss2-D maps failed to yield any certain temporal or location results.
PDB had no structure matches for YGR201C.
PUMA2 had no information pertaining to my gene.
YAC Two-hybrid analysis of protein interaction also failed to yield results.
DIP was a useful analysis tool.

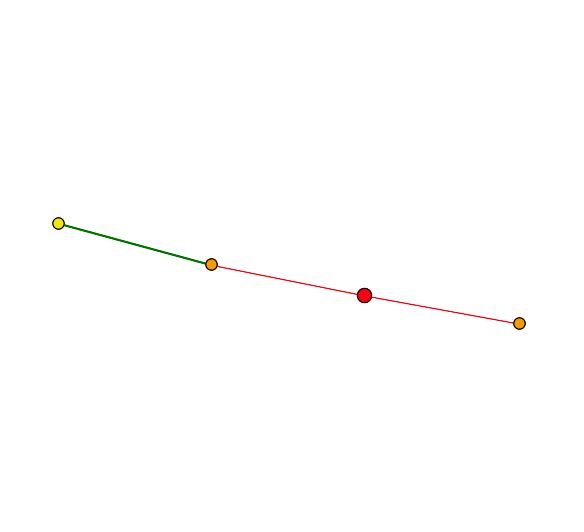
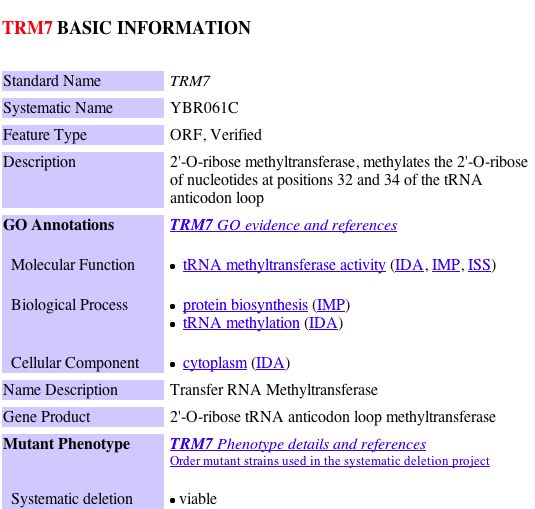
| The primary connections for YGR201C are to a probable membrane protein YBR077C and Trm7p which is involved in t-RNA metabolism and protein biosynthesis in the cytoplasm. The second shell node is YKR007W which has an unknown function but can be found in the vacuolar membrane. YGR201C appears to be related to membrane proteins. Both of the primary connections are a result of high-throughput analysis and have not be confirmed experimentally. The figures below were taken from SGD. |
 |
 |
MIPS
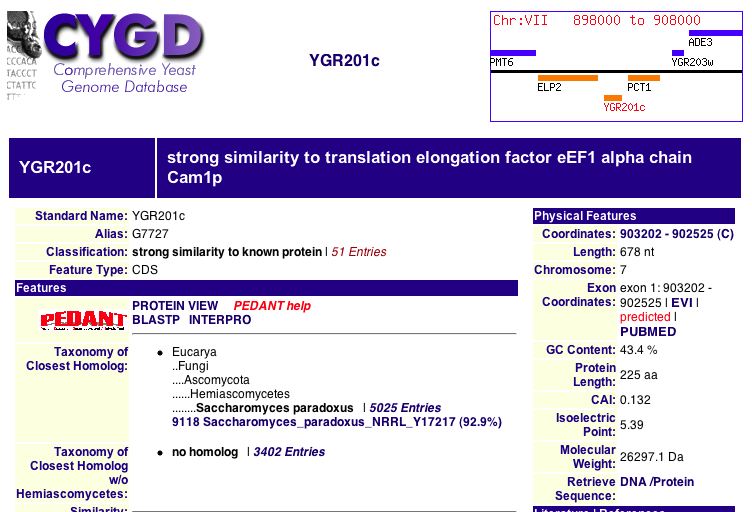
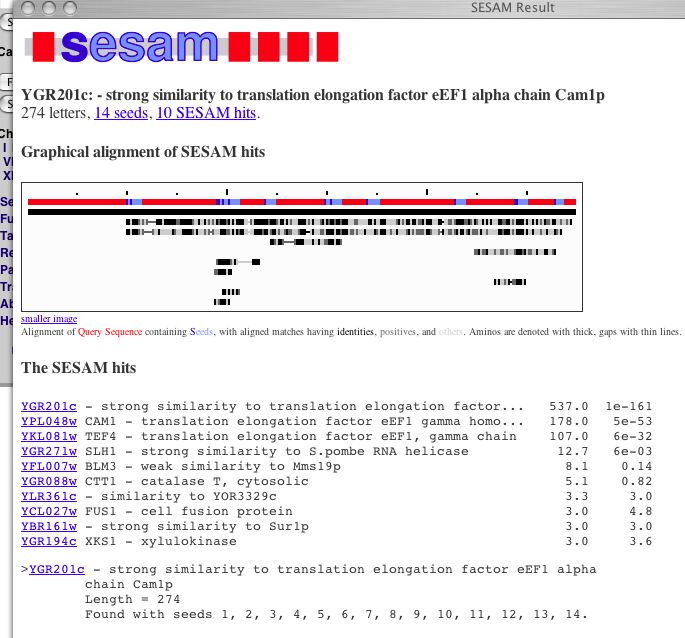
| This is the basic information from the MIPS page. The protein has an isoelectric point of 5.4 and a molecular weight of 26297 Da. This figure identifies YGR201C as a homolog to proteins responsible for translation elongation. Another part of MIPS is SESAM (Seed Extraction Sequence Analysis Method), a method of comparing amino acid sequences to find paralogs. |
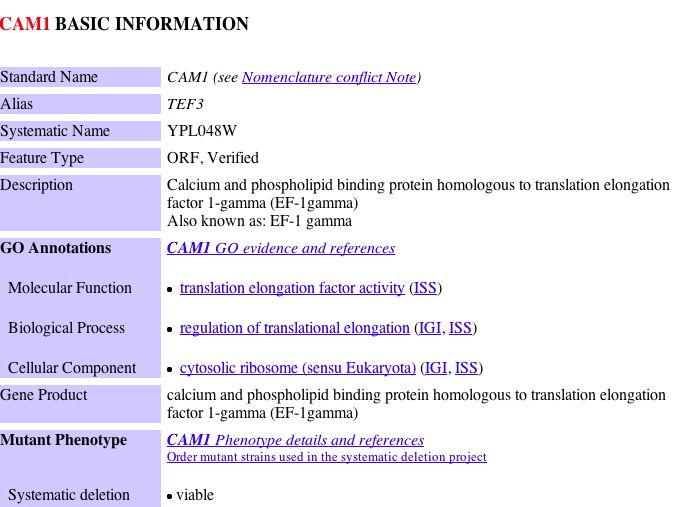 |
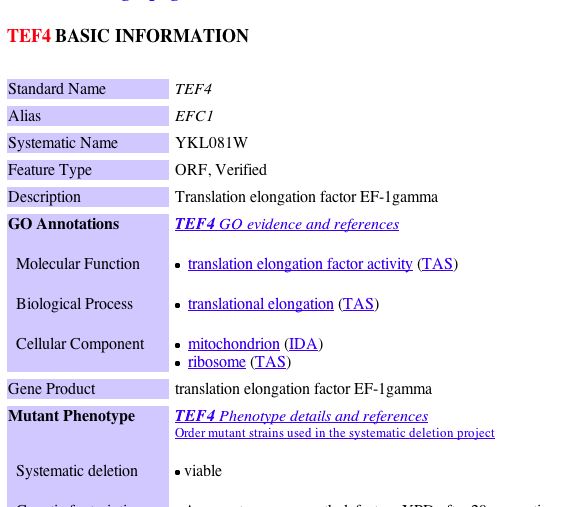 |
 |
Conclusions and Experiments: I believe that YGR201C is involved in translation elongation or transport after this study. A limited amount of interaction data was available; however, the data that I found seemed to provide information leading to similar conclusions. To better understand the function of YGR201C, I would like to plan a knockout experiment and observe the effects on translation efficiency. It is possible that this protein would have a homolog that could take over when my ORF was gone so translation could continue without interruption. If so, additional experiments with more than one gene knocked out would be informative. Another potential protein function is transport of materials, particularly those required for translation. A localization experiment would be helpful in trying to uncover YGR201C's function. Fluorescent antibody tags would be ideal for labeling this protein. |
References
Dove, Alan. (1999) Proteomics: translating genomics into products? Nature Biotech. 17: 233-236.
Campbell AM, Heyer LJ. 2003. Discovering Genomics, Proteomics, and Bioinformatics. Benjamin Cummings: San Francisco. p.162-204.
Kumar, A., Cheung, K.-H., Ross-Macdonald, P., Coelho, P.S.R., Miller, P., and Snyder, M. (2000). TRIPLES: a Database of Gene Function in S. cerevisiae. Nucleic Acids Res. 28, 81-84. (Full-text in PDF reproduced with permission from NAR Online http://www.oup.co.uk/nar )
Ross-Macdonald, P., Coelho, P.S.R., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.-H., Sheehan, A., Symoniatis, D., Umansky, L., Heidtman, M., Nelson, K., Iwasaki, H., Hager, K., Gerstein, M., Miller, P., Roeder, G.S., and Snyder, M. (1999). Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402, 413-418. Nature 402, 413-418.
| MIPS: CYGD |
| PDB |
| PUMA2 |
| Swiss-2DPAGE |
| TRIPLES |
Questions or Comments? Email: shbossie@davidson.edu
© Copyright 2004 Department of Biology, Davidson College, Davidson, NC 28036