This web page was produced as an assignment for an undergraduate course at Davidson College.
SEC22/YLR268W and its Non-annotated Neighbor YLR264C-A
SEC22 is a well annotated Saccharomyces cerevisae gene on chromosome 12 that encodes a synaptobrevin homolog (v-SNARE protein). The v-SNARE protein is involved in anterograde and retrograde transport between the ER and the Golgi. While SEC22 is well-annotated, its nearby neighbor, YLR264C-A, is not (Dolinski et. al., 2004).
Chromosomal Location:

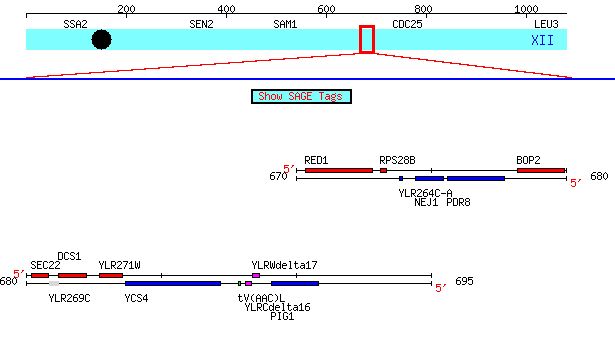
Fig 1. Visualization of proximity of YLR264C-A and SEC22 on chromosome XII of Saccharomyces genome (coordinates: 670202-690846). Image from: (Cherry et. al., 1997; <http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000004258>). Permission Granted.
SEC22 BASIC GENE INFORMATON:
Saccharomyces Genome Database: Links from the SGD database can lead us through a web of information regarding nearly all significant aspects of the gene. SGD informs us that the null mutant, although viable, is cold and heat sensitive as well as defective in ER to Golgi transport. SGD describes SEC22 as a gene that encodes a v-SNARE protein that is found on ER and Golgi membranes and plays a role in anterograde and retrograde transport between the ER and Golgi. The v-SNARE protein is a synaptobrevin homolog (Dolinski et. al., 2004; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SEC22>). Synaptobrevin is a common vesicle-associated membrane protein (NCBI, 2004; <http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=51704193>) Synaptobrevin homologs are found in mammals such as mice where these proteins regulate membrane traffic between the ER and the Golgi (NCBI, 2004; <http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=48474741>). Thus, an improved understanding of the yeast genome has potential to benefit multiple fields.
MOLECULAR FUNCTION:
Concisely, the Gene Ontology molecular function of SEC22 is v-SNARE activity. v-SNARE proteins, of which S. Cerevisae contains 14, catalyze fusion events between intracellular membranes (Dolinski et. al., 2004; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SEC22> and Dolinski et. al., 2004;<http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=5485#is>). SEC22 has a 3D shape that allows it to bind specifically to proteins on the membrane of Golgi and catalyse fusion events of the ER and the Golgi, as well as among the stacks of Golgi.
According to Gene Ontology, the molecular function of SEC22 protein is v-SNARE activity (GO:0005485). v-SNARE activity is less accessible to immediate understanding than some more frequently discussed molecular functions such as ATPase activity; however, v-SNARE acitivty is easy to understand. Yeast contains 14 v-SNARE proteins whose molecular functions encompass specific membrane fusion events that ensure specificity in vesicle targeting and secretory pathways. The SEC protein in question, SEC22, is able to direct traffic from the Golgi to the ER, and vice versa, because the SEC22 protein binds to specific proteins on the ER membrane. In simple terms, the molecular function of SEC22 lies in, derives from, or is dictated by its 3D structure because the specific shape of SEC22 confers the ability to selectively bind to other membrane proteins (Gene Ontology, 2004; <http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=5485#amigo>).
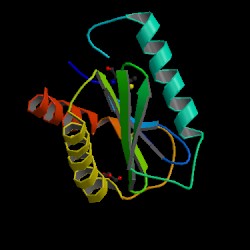
Fig 2. PDB image of an engineered SEC22 homolog obtained from Mus Musculus, 1ifq, can be viewed at the adress in the follwing citation (PDB, 2004; <http://pdbbeta.rcsb.org/pdb/explore.do?structureId=1ifq>). However, Note that the amino acid sequence is only 40% identical and 35% similar (see Fig 3). Also note that the SEC22B Vesicle Trafficking protein is only 127 aa's long whereas the S. Cerevisae SEC22 protein is 215aa's long (Dolinski et.al., 2004; <http://db.yeastgenome.org/cgi-bin/protein/get3d?locus=YLR268W&pdb=1ifq_B&align=1>) . Other views of the protein are offered at the PDB site ([PDB], 2004; <http://www.rcsb.org/pdb/cgi/explore.cgi?job=graphics;pdbId=1ifq&bio=1&opt=show&size=250#>).
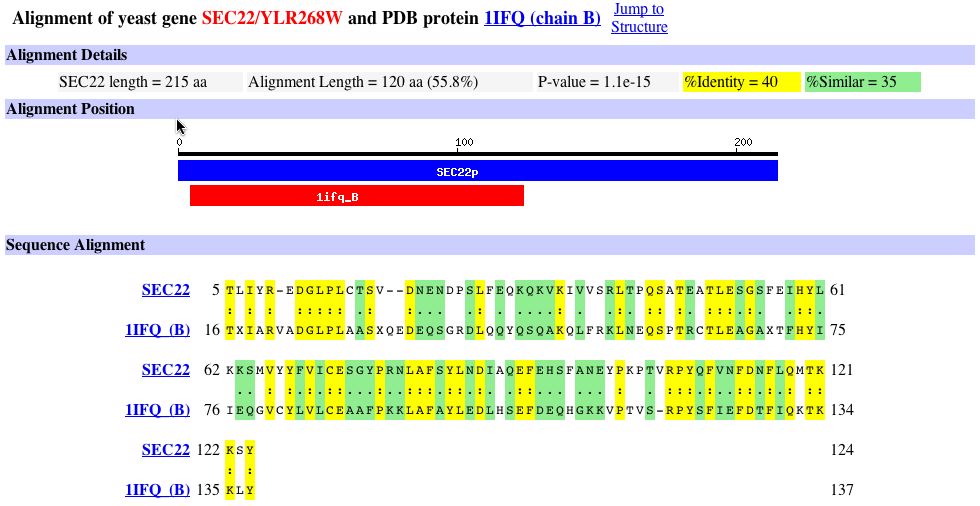
Fig 3. The top of the figure, under the section alignment details, indicates that the amino acid sequence is only 40% identical and 35% similar (Dolinski et.al., 2004; <http://db.yeastgenome.org/cgi-bin/protein/get3d?locus=YLR268W&pdb=1ifq_B&align=1>).
BIOLOGICAL PROCESS:
Concisely, SEC22 contributes to the biological process of intracellular protein targetting by ensuring appropriate and specific membrane fusion events between the ER and the Golgi.P Proteins must be localized to the appropriate places in the cell in order for these proteins to be functional. Remember Cystic Fibrosis. SEC22 contributes to the important process of localizing proteins. SEC22 may contribute to timing of cellular responses because some proteins are must be localized only in times of stress. (Burri et. al., 2004; <http://www.blackwell-synergy.com/links/doi/10.1046/j.1600-0854.2003.00151.x/full/>).
Four Gene Ontology biological processes are listed for SEC22 on the SGD page including: ER to Golgi transport; intra-Golgi transport; Golgi to ER retrograde transport; and vesicle fusion. ER to Golgi transport is defined by gene ontology as follows: "the directed movement of substances from the endoplasmic reticulum (ER) to the Golgi, mediated by COP II vesicles. Small COP II coated vesicles form from the ER and then fuse directly with the cis-Golgi. Larger structures are transported along microtubules to the cis-Golgi." Gene ontolog defines Intra-Golgi transport as "the directed movement of substances within the Golgi, mediated by small transport vesicles. These either fuse with the cis-Golgi or with each other to form the membrane stacks known as the cis-Golgi reticulum (network)." Retrograde transport is the "The directed movement of substances from the Golgi back to the endoplasmic reticulum, mediated by COP I vesicles." Finally, vesicle fusion, if we need a definition, is "fusion of the membrane of a transport vesicle with its target membrane." We might expect that a protein with so many roles is essential to function; however, there is likely some overlap with other proteins because the null mutant is viable, although it does suffer increased sensitivity to heat and cold as well as decreased effectiveness of transport from the ER to the Golgi (Dolinski et. al., 2004; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SEC22>). We must remember that the number of biological processes listed is not necessarily related to the necessity of the protein. More isn't always better.
The numerous listed functions of the SEC22 protein reflect the breadth, but also the limited nature of our knowledge about SNARE family. We know that SEC22 uses mediates specific vesicle fusion in order to direct the transport of proteins from the Golgi to the ER , the ER to the Golgi, and from one stack of the Golgi to another stack of the Golgi. At the same time, there is a great deal that we do not know about SNARE proteins , such as the mechanisms by which SNARE proteins (e.g. SEC22) perform their roles and themselves get targetted to appropriate cellular locations (NCBI, 2004; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15123693>). In January 2004, Lenna Burri and Trevor Lithgow compiled an article summarizing our current state of knowledge regarding SNARE proteins (Burri et. al., 2004; <http://www.blackwell-synergy.com/links/doi/10.1046/j.1600-0854.2003.00151.x/full/>)
CELLULAR COMPONENT:
SGD defines three regions of localization for SEC22: Golgi apparatus; endoplasmic reticulum; and integral to membrane. Burri et. al., (2004) specified that SEC22 is a type II membrane protein: that is, it has a a C-terminal segment of polypeptide that serves as a membrane anchor. Twenty of the twenty four SNARE proteins appeared to be type II membrane proteins. SEC22 appears to be one of only four SNARE proteins that help dictate the transport of other SNARE proteins between compartments because SEC22 contains a cytoplasmic sorting sequence that can directly interact with coat proteins on other membranes. Burri et. al. (2004) has a stockpile of helpful images.
UNKNOWN GENE: YLR264C-A
NUCLEOTIDE SEQUENCE INFORMATION:
Since nothing is known about the gene I started with the proposed mRNA nucleotide sequence (See Fig 5) and the amino acid sequence.
Sequence Accession number: U17244. YLR264C-A on chromosome XII from coordinates 673946 to 673830.
ATGAATGTAATATTTAAATTGTATTTAACGATAGATGCACCAAAAAAAAAGAAAGTTACAGTAAGAGACTTCTTGTCGATTCGTTATTCCTGCCATACAGGTTAGCCTCAAATTAA YLR264C-A mRNA/cDNA coding sequence (Dolinski et. al., 2004;<http://db.yeastgenome.org/cgi-bin/getSeq?map=nmap&seq=YLR264C-A&flankl=&flankr=&rev=>)
AMINO ACID SEQUENCE INFORMATION: 38aa's
Used BLASTx and the SGD SEC22 page links to obtain convergent data regarding the trnaslated sequence. The sequence can be found at the bottom of the following webpage
(NCBI, 2004; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=Protein&list_uids=33438841&dopt=GenPept>).
Accession Number: NP_878129; RefSeq Accession#: NC_001144.3<
Translated Sequence: MNVIFKLYLTIDAPKKKKVTVRDFLSIRYSMPYRLASN
CONSERVED DOMAIN SEARCHES:
I performed a conserved domain search on NCBI and returned zero hits.
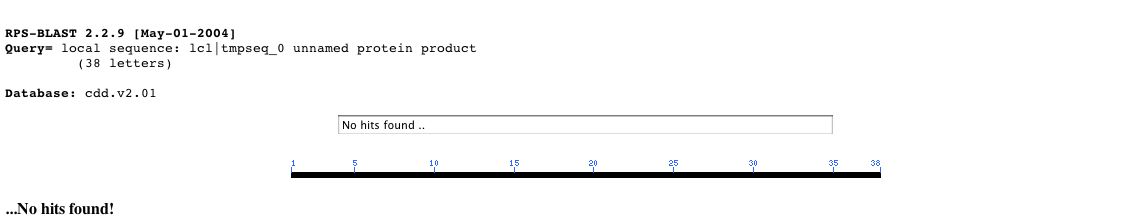
Fig 4. NCBI COD search returned no hits. To achieve the above photograph one must enter the protein sequence listed in the sequence information section (i.e. MNVIFKLYLTIDAPKKKKVTVRDFLSIRYSMPYRLASN) into the COD search page at the following link (<Marchler-Bauer et. al., 2003; <http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi>)
I found no homologs on the mips (Munich Information Center for Protein Sequences) page for the YRL264C-A (MIPS, 2004; < http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YLR264C-A>). However, I did discover that the GC content of the region is 29.9%, which is interesting considering what we learned earlier in the year about the correlation between GC content and coding regions.
I performed a PSI-BLAST search and did not return significant alignments other than the one with the gene under investigation.

Fig 5. PSI BLAST did not return any potential conserved domains. The PSI-BLAST hompage can be found at the following citation (NCBI, 2004; <http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi>). One may enter the protein sequence on this page to obtain the same results as those displayed in the picture above.
STRUCTURAL ANALYSIS:
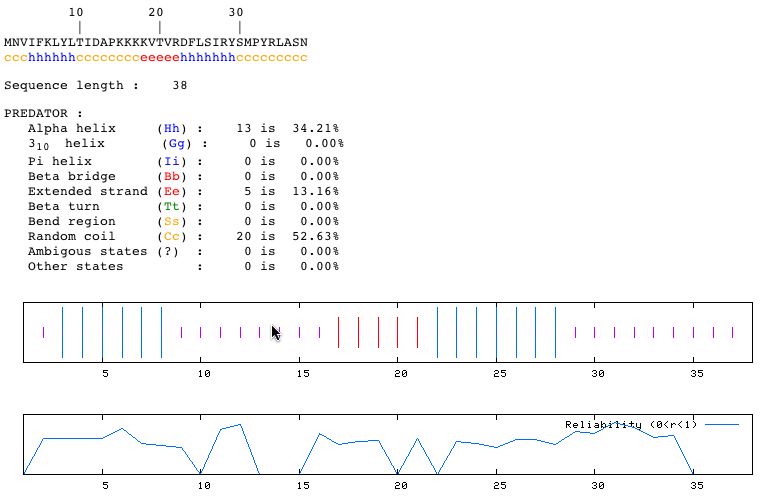
Fig 6. PREDATOR Secondary Structure Analysis: PREDATOR returned results that predict: 2 alpha helices of 5 and 7aa's each; 3 random coils of 3, 8, and 9aa's each; and a 5aa extended strand for our 38aa protein. One can paste the amino acid sequence to the PREDATOR homepage to get the above results (PREDATOR, 2004; <http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html>).
KYTE-DOOLITTLE ANALYSIS:
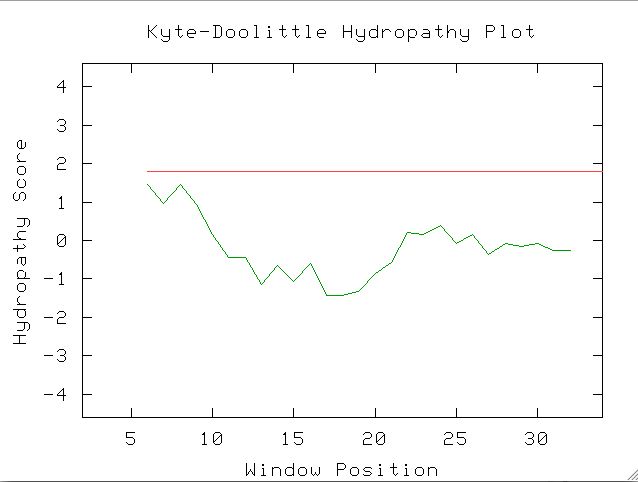
Fig 7. A Kyte-Doolittle Plot of window size 11 does not indicate transmembrane domains. Regions above the red line are hydrophobic, and highly negative regions are hydrophilic. One can enter the protein sequence information at the following page (Kyte-Doolittle, 2004; <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/kyte-doolittle.htm>)
PDB ANALYSIS:

Fig 8. PDB homologous were not found. Image adapted from (Dolinski et.al., 2004; <http://db.yeastgenome.org/cgi-bin/protein/get3d?sgdid=S000028808>).
CONCLUSION: Due to the dearth of information on the sequence and the fact that it is exceedingly small (i.e. 38 aa's), combined with the fact that SGD lists the feature type of this ORF as "ORF, Uncharcterized" I must preliminarily conclude that this ORF is actually an exon that is part of a larger nearby protein (Dolinski et. al., 2004 <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=YLR264C-A>). If we refer to the map at the top of the page we can see that nearby ORFs on the bottom strand include NEJ1 and PDR8 (Dolinski et. al., 2004; <http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000004258>). Without further data it is difficult to say anything except that it is possible that YLR264C-A is involved in the alternative splicing of nearby ORFs NEJ1 or PDR8; however, this conclusion is more akin to a hypothesis than a conclusion because it is highly speculative.
REFERENCES:
Burri et. al., 2004. A Complete Set of SNAREs in Yeast. Traffic 5: p.45 <http://www.blackwell-synergy.com/links/doi/10.1046/j.1600-0854.2003.00151.x/full> Accessed October 5. 2004
Cherry JM. et. al., 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67-73.
Dolinski, K. et. al., 2004. Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed Oct. 4, 2004.
Gene Ontology Software Group, 2004. Gene Ontology Database.<http://www.godatabase.org/dev/database/> Accessed Oct. 4, 2004.
Kyte J and Doolittle R, 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105-132.
Marchler-Bauer et. al., 2003; "CDD: a curated Entrez database of conserved domain alignments", Nucleic Acids Res. 31:383-387. < http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi> Accessed Oct. 5, 2004
[MIPS] Munich Information Center for Protein Sequences, 2004. <http://mips.gsf.de/> Accessed Oct. 5, 2004
[NCBI] National Center for Biotechnology Information, 2004. <http://www.ncbi.nih.gov/> Accessed Oct. 4, 2004.
[PDB] Protein Database, 2004.<http://www.rcsb.org/pdb/> Accessed Oct. 5, 2004.
[PREDATOR] Pôle Bioinformatique Lyonnais, 2004. <http://pbil.ibcp.fr/html/pbil_index.html> Accessed Oct. 5, 2004.
LINKS:
Genomics Page
Biology Home Page
E-mail Questions and Comments.