This web page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Gene Expression:
On my first Favorite Gene Page I used online genome databases to describe one annotated gene SEC22 and one of its non-annotated neighbors, YLR264C-A. On this page my goal is to enhance my previous characterization of the genes by using public microarray databases. These databases contain the results of microarray experiemtns. The data is public because of the magnitude of the data is unprecedented. Microarray experiments measure simultaneously the expression levels of all genes in a genome. By correlating the expression patterns of unkown genes with known genes we can come up with new hypotheses about the function of nonannotated genes and obtain evidence for the function of annotated genes. To learn more about microarray methodology check out the following link (DNA Microarray Methodology - Flash Animation, 2004; <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/method/chip/chip.html>).
Annotated Gene - SEC22
The gene SEC22 is located on chromosome 12 in Saccharomyces cerevisae. It belongs to a family of 14 genes in yeast that have v-SNARE activity. In order to better orient yourself with SEC22, its function and v-SNARE activity see my Favorite Yeast Gene page. Here, I will offer a brief description in terms of gene ontology:
Biological Process- Concisely, SEC22 contributes to the biological process of intracellular protein targetting by ensuring appropriate and specific membrane fusion events between the ER and the Golgi.P Proteins must be localized to the appropriate places in the cell in order for these proteins to be functional. Remember Cystic Fibrosis. SEC22 contributes to the important process of localizing proteins. SEC22 may contribute to timing of cellular responses because some proteins are must be localized only in times of stress. (Burri et. al., 2004; <http://www.blackwell-synergy.com/links/doi/10.1046/j.1600-0854.2003.00151.x/full/>).
Molecular Function- Concisely, the Gene Ontology molecular function of SEC22 is v-SNARE activity. v-SNARE proteins, of which S. Cerevisae contains 14, catalyze fusion events between intracellular membranes (Dolinski et. al., 2004; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=SEC22> and Dolinski et. al., 2004;<http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=5485#is>). SEC22 has a 3D shape that allows it to bind specifically to proteins on the membrane of Golgi and catalyse fusion events of the ER and the Golgi, as well as among the stacks of Golgi.
Cellular Component- SEC22 is an integral membrane protein involved in specific membrane fusions events. SEC22 can be found in two membranes: Golgi apparatus; endoplasmic reticulum.
The source of the following microarray results should first be noted. All of the following images were obtained from the Expression Connection (Expression Connection, 2004, <http://db.yeastgenome.org/cgi-bin/SGD/expression/expressionConnection.pl>). In examining the different microarrays that were available, I attempted to select for those that suggested something of particular interest with regards to SEC22 expression.
Fig 1. This is the color scale employed in pictures from Expression Connection. Red indicates gene induction in the experimental condition. Green indicates gene repression in the experimental condition.
Similarly Expressed Genes: Expression during the diauxic shift DeRisi JL, et al.
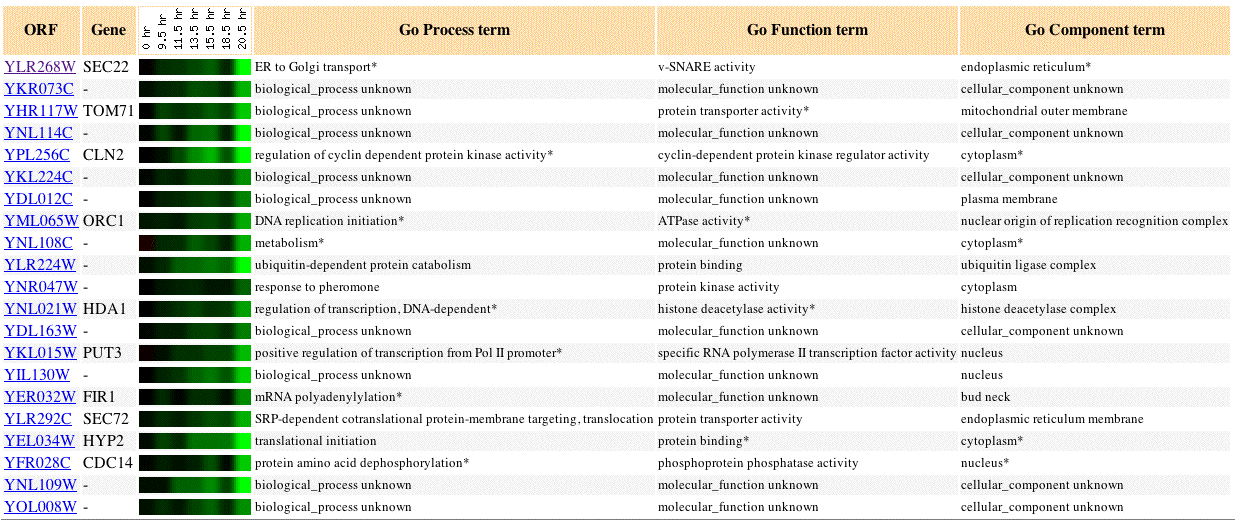
Fig 2. Image of SEC22 expression during diauxic shift, as well as, 20 similar genes (Pearson's correlation coefficient, p>0.8).
Diauxic shift in yeast is the change in respiration type from anaerobic to aerobic. A change in respiration type causes different genes to become induced or repressed. In this experiment, SEC22 is repressed. SEC22 shares a similar repression profile to other membrane tagetting and translational regulation genes: SEC72, HDA1, PUT3, HYP2. The SEC72 protein is especially similar in expression profile to SEC22. Both SEC22 and SEC72 exhibit an increase in the intensity of induction from 9.5 to 20.5 hours.
Similarly Expressed Genes: Expression in response to histone depletion Wyrick JJ, et. al.
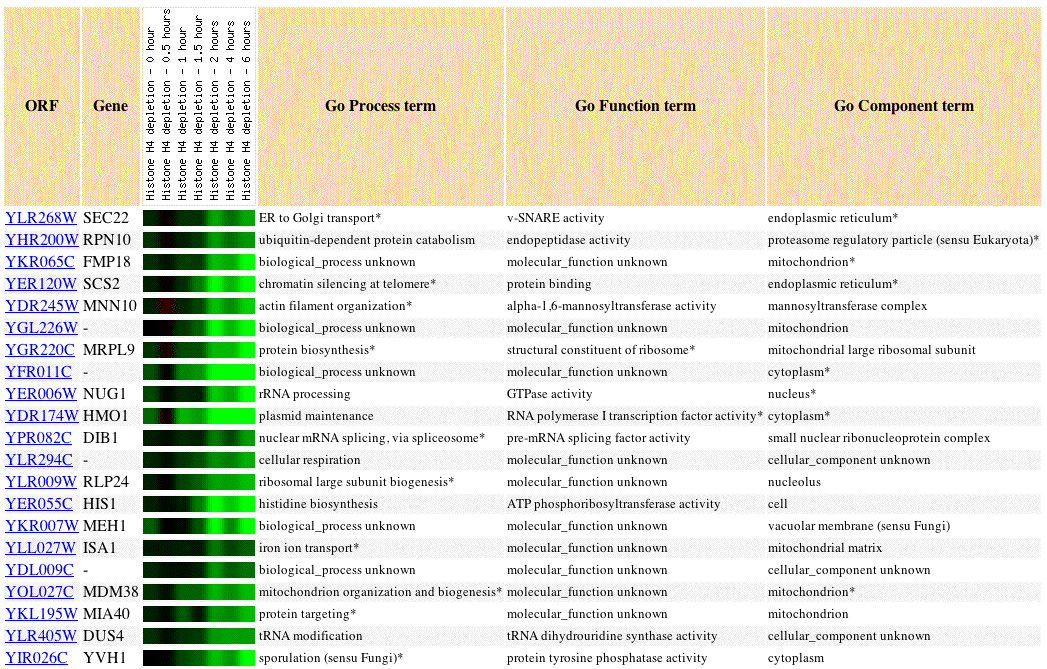
Fig 3. Image of SEC22 expression profile in response to histone depletion, as well as the expression profiles of 20 similar genes (Pearson's correlation coefficient, p>0.8).
Histones are proteins that package DNA into nucleosomes. In the above experiment histones were allowed to be depleted in the experimental condition. Loss of histones caused repression of SEC22. Other genes involved in protein/tRNA targetting and transport showed similar expression profiles to SEC22: MIA40, DIB1, NUG1, DUS4. This is consistent with our knowledge of SEC22 as a gene involved in fusion events of the ER membrane because protein transport through the ER is closely tied to post-translational modification.
Similarly Expressed Genes: Expression during the unfolded protein response Travers et. al.
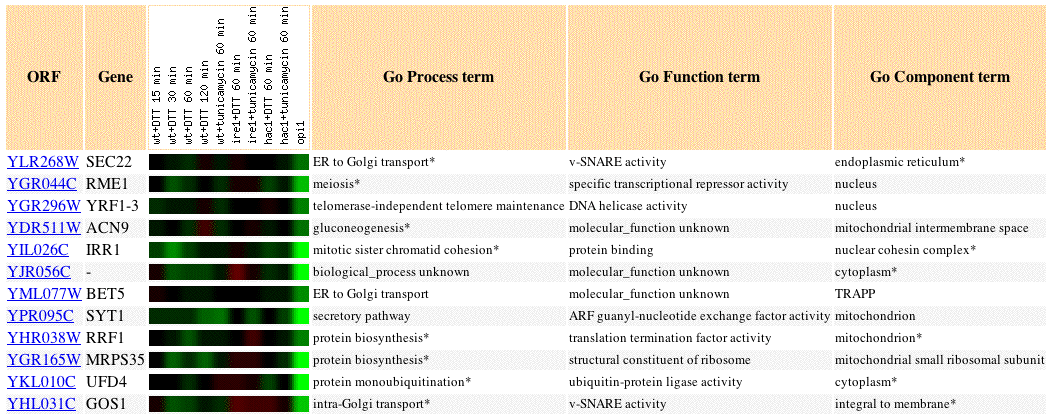
Fig 4. Image of SEC22 expression profile in response to stress placed on the endoplasmic reticulum, as well as the expression profiles of 11 similar genes (Pearson's correlation coefficient, p>0.8). The highest greatest amount of repression in this experiment was 1.6 fold repression.
The unfolded protein response (UPR) is the coordinated induction or repression of genes in response to Endoplasmic Reticulum (ER) stress. A study that investigates UPR is expecially important because SEC22 is supposedly involved in ER to Golgi transport. The above figure illustrates genes with expression profiles similar to SEC22 that are involved in translation and secretory pathways: BET5, GOS1, SYT1, RRF1. The fact that genes with similar GO Processes have similar expression profiles supports the guilt by association hypothesis for this data set and supports current knowledge of SEC22 function.
Similarly Expressed Genes: Expression in response to varying zinc levels Lyons et. al.
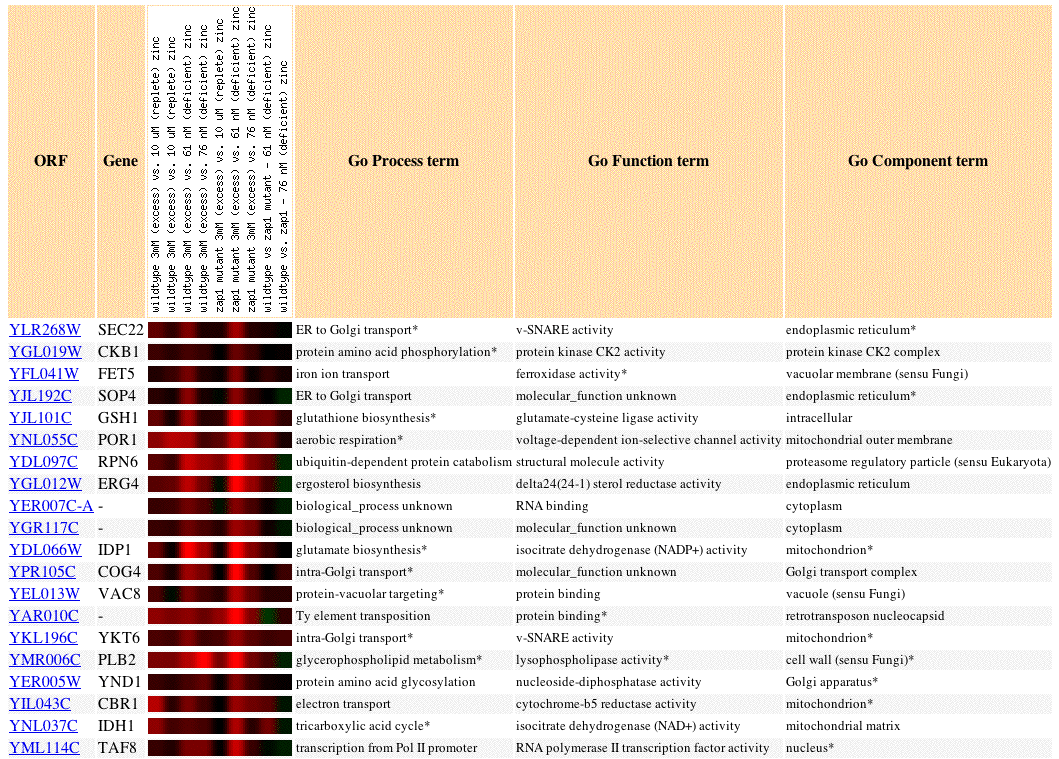
Fig 5. Image of SEC22 expression profile in response zinc concentration levels, as well as the expression profiles of 19 similar genes (Pearson's correlation coefficient, p>0.8).
Genes with expression profiles similar to SEC22 that are involved in translation and secretory pathways include: SOP4, FET5, COG4, and YKT6. If we accept the guilt by association hypothesis then this convergent data supports current knowledge of SEC22 as a protein involved in ER to Golgi transport. Sometimes, apparently unrelated genes share expression patterns. It is instructive to resolve these discrepancies when possible because we may form new hypotheses and recognize flaws in our current methods
ERG4 resembles v-SNARE proteins in its expression profile. ERG4 is involved in ergosterol biosynthesis. We know that steroid synthesis occurs in the SER. Thus, it would be reasonable to expect that SER function would be slowed as ER function is slowed. We grow accustomed to thinking that genes transcribe proteins. We tend to forget about steroids and lipids? We should guess that steroid synthesis gets slowed when ER secretory pathways slow down. This provides an example of how flexible we must be when coming up with hypotheses for gene functions and cellular locations of non-annotated genes. The guilt by association hypothesis has limits--results can be interpreted in different ways.
Conclusion
The guilt by association hypothesis (i.e. that genes with similar expression profiles are often related in GO function or process) is supported by the above data and supports our knowledge of SEC22 function. We saw that genes involved in Golgi transport and post-translational modification often get expressed similarly in response to environmental stress. Genes with functions similar to SEC22 often have simialr expression profiles. The nature of discovery science and all good science and knowledge is that we remain aware of our assumptions, such as the guilt by association hypothesis.
YLR262C-A
On this webpage I endeavored to hypothesize the function of YLR264C-A. Microarray databases represent the best tools we have for coming up with hypotheses about the functions of non-annotated genes. As such, I mined public databases of microarray data. My goal was to correlate the expression patterns of YLR264C-A with the expression patterns of genes with known functions. I did not find any expression data on my unknown gene YLR264C-A, so I analyzed expression patterns for a nearby non-annotated gene, YLR262C-A.
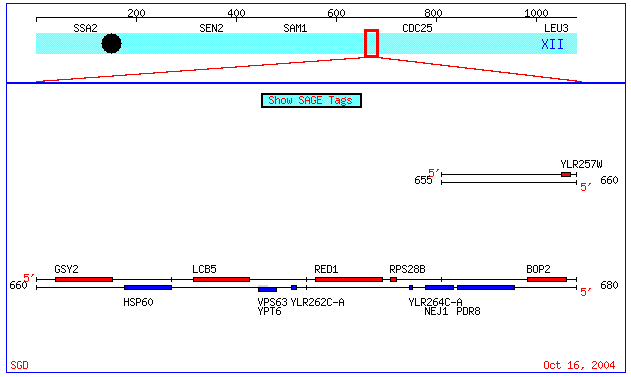
Fig 6. Visualization of proximity of YLR264C-A and YLR262C-A on Chromosome XII of the Saccharomyces genome (coordinates: 659470 - 679664). Image from: (Cherry et. al., 1997; <SGD Picture>). Permission Granted.
I began my search at Expression Connection:
Similarly Expressed Genes: Gene regulation by Swr1, Htz1, and Ino80 Mizuguchi G, et al.
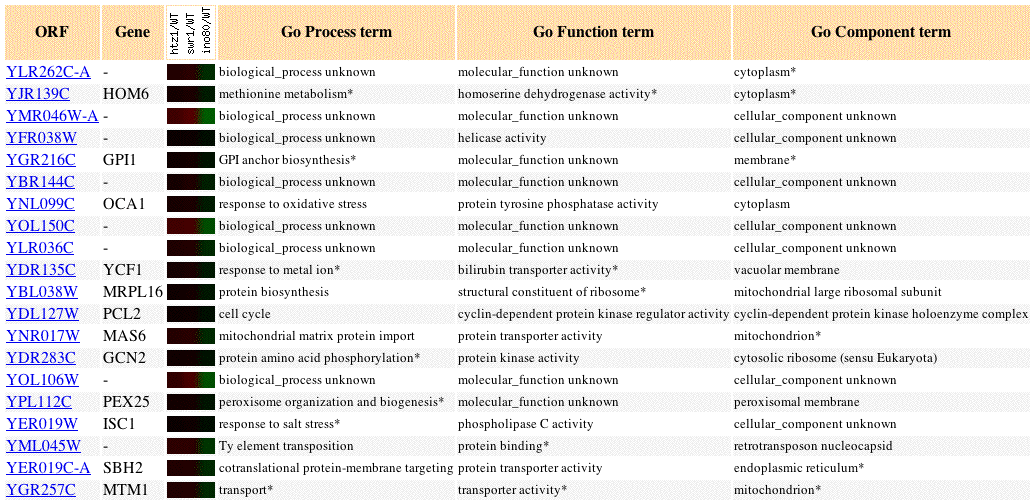
Fig 7. Image of YLR262C-A expression profile in response to gene regulation by as well as the expression profiles of 19 similar genes (Pearson's correlation coefficient, p>0.8). Mizuguchi et. al. was the only paper that returned a returned a result for YLR262C-A. Image adapted from: (SGD, 2004; <http://db.yeastgenome.org/cgi-bin/expression/expressionConnection.pl?orf=YLR262C-A&dataset=swr1_htz1_ino80&type=similar>). Obtain my search result by visiting the search page listed at the end of the paragraph. Type YLR262C-A into the search box, and mark all the papers. (SGD, 2004 <http://db.yeastgenome.org/cgi-bin/expression/expressionConnection.pl>)
Mizuguchi et. al. was the only paper, out of 16, that returned a result. Many of the genes in the above figure have unkown functions, yet, the above figure gives us some leads.
We know the biological process of some of the genes. HOM6 perfoms methionine metabolism. Many of the genes are involved in transport: MAS6, SBH2, MTM1, YCF1. Many of the genes are involved in detoxification: PEX25, HOM6, YCF1. The function junction search also returned detoxification genes.
Function Junction:
Microarray Global Viewer
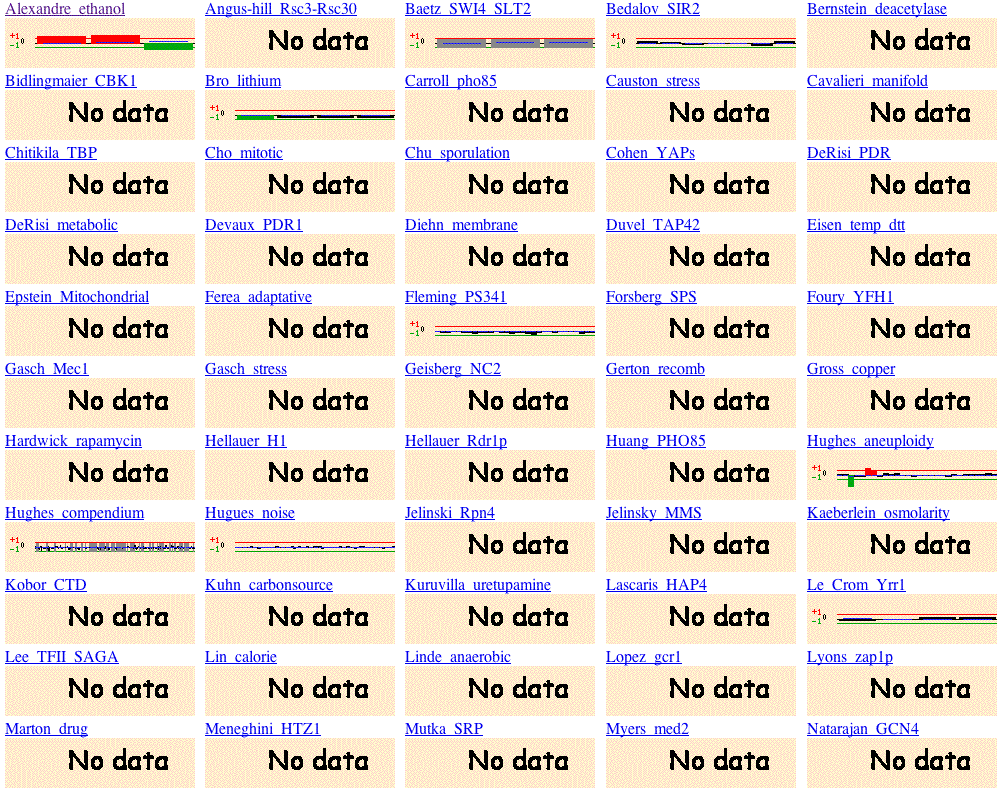
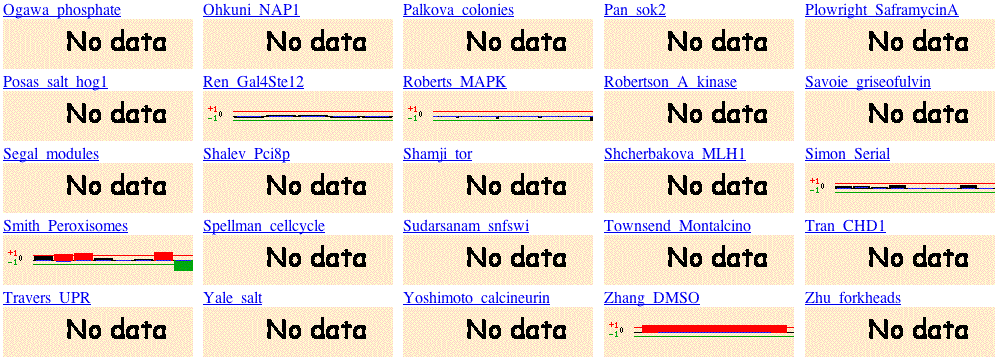
Fig 8. Microarray Global viewer obtained from Function Junction search. To duplicate these results, type YLR262C-A into the following search site and scroll down the page until you see the heading, microarray global viewer (Function Junction, 2004; <http://db.yeastgenome.org/cgi-bin/SGD/functionJunction>).
The Microarray Global viewer, although short on data, allows us to hypothesize about the function of the gene. YLR262C-a is induced in the experiments that involve a response to toxicity. YLR262C-A is induced in Alexandre's ethanol experiement, Smith's peroxisome experiment and Zhang's DMSO experiment. We must preliminarily conclude that the gene is involved in detoxification.
Conclusion
YLR262C-A appears to be involved in cellular detoxification. The images from expression connection and the one from function junction support this hypothesis.
I would like to know more about the relation between SEC22 and YLR262C-A. Do they have related functions that remain hidden due to the dearth of data on YLR262C-A. Do their shared expression patterns remain hidden? Some data suggests a link between these two genes. The expression connection analysis for YLR262C-A showed a number of genes involved in protein binding and transport: MAS6, SBH2, MTM1, YCF1, YML045W. SEC22 also shared expression patterns with genes related to protein binding, and transport. Unique and unexpected similarities include the presence of Ty element proteins in Mizuguchi et al. (2004) and Lyons et. al. (2000). Before we get carried away searching for every similarity, we should remind ourselves that any similarity between SEC22 and YLR262C-A is speculative at this time due in part to the limited data.
We do not have enough data to say anything about YLR262C-A except that YLR262C-A is likely involved in detoxification.
References:
Burri et. al., 2004. A Complete Set of SNAREs in Yeast. Traffic 5: p.45 <http://www.blackwell-synergy.com/links/doi/10.1046/j.1600-0854.2003.00151.x/full> Accessed October 5. 2004.
Cherry JM. et. al., 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67-73.
DeRisi et. al., 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-6.
Dolinski, K. et. al., 2004. Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed Oct. 4, 2004.
DNA Microarray Methodology - Flash Animation, 2004; <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/method/chip/chip.html> Accessed October 17, 2004.
Expression Connection, 2004, <http://db.yeastgenome.org/cgi-bin/SGD/expression/expressionConnection.pl>. Accessed October 17, 2004.
Function Junction, 2004; <http://db.yeastgenome.org/cgi-bin/SGD/functionJunction> Accessed October 18, 2004.
Lyons et. al., 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci U S A 97: 7957-62.
Mizuguchi et. al., 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343-8.
Travers et. al., 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249-58.
Wyrick et. al., 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418-21.
LINKS:
Genomics Homepage
Biology Homepage
Max Citrin's Genomic Homepage
E-mail Questions and Comments
© Copyright 2004 Department of Biology, Davidson College, Davidson, NC 28035