This web page was produced as an assignment for an undergraduate course at Davidson College
Analysis of the Expression Patterns of Cox17 and the Non-annotated YLL007C Yeast Genes
|
Summary of Previous Work:
Cox17:
Cox17 is involved in cytochrome c oxidase biogenesis and intra-cellular copper ion transport. Cox17 has been shown to be essential for the expression of cytochrome oxidase in Saccharomyces cerevisiae (Beers et al. 1997). Cox17 is a soluble protein found in the cytoplasm and intermembrace space of the mitochondria in yeast. Yeast cells that harbor a mutant Cox17 gene are respiratory-deficient but are still able to synthesize mitochondially and nuclearly encoded cytochrome oxidase units. This respiratory defect can be partially rescued by high exogenous Cu(II) levels, providing further evidence that Cox17 in involved with copper ion delivery to the mitochondria (Heaton et al. 2000).
YLL007C:
YLL007C is considered to be a hypothetical ORF on chromosome XII of S. Cerevisiae with no known molecular function or cellular process. study conducted by Giaever G, et al. in 2002 indicated that mutant yeast with a systemic deletion of YLL007C were still viable. However, mutations that result in null YLL007C alleles along with null alleles of pac10, gim4, or gim5 resulted in a slow growth phenotype (Tong et al. 2004). Another study using genetic footprinting analysis of the S. Cerevisiae genome used the Ty1 transposon as a mutagenizing agent. Mutations involving the YLL007C ORF resulted in several apparent "severe growth defects" after 20 generations (Dunn et. al 2004). Based upon Kyte-Doolittle Hydropathy Plot Analysis, I predicted that YLL007C is a transmembrance protein. Since homoolgy and sequence data yielded no conclusive results, I based my previous predictions regarding the cellular role of YLL007C on studies involving mutant forms of the YLL007C gene. I hypothesized that the S. Cerevisiae YLL007C gene is involved in cellular growth, thus potentially playing a role in the regulation of the cell cycle. It should also be noted that PSI-Blast analysis of YLL007C indicated that the gene had some similarity to human and murine cell motility and engulfment proteins. Although this was not discussed in great detail in the previous assignment, I have recently determined that the human protein encoded by this gene, called EMLO1, interacts with the dedicator of cyto-kinesis 1 protein to promote phagocytosis and effect cell shape changes. Similarity to a C. elegans protein suggests that this protein may function in apoptosis and in cell migration.component of signalling pathways that regulate phagocytosis and cell migration and is the mammalian orthologue of the C. elegans gene, ced-12. CED-12 is required for the engulfment of dying cells and cell migration (Enterez Gene). Since the yeast YLL0076 protein has approximately 65 percent alignment with this human protein, it is possible that the YLL007C protein could have a similar function. |
Analysis of the Cox17 Expression Pattern:
Expression During the Cell Cycle:
In a study conducted by Spellman et al. in 1998, those genes whose transcript levels varied periodically during the cell cycle were identified via microarray hybridization. It is hypothesized that such fluctuation during the cell cycle might be necessary to maintain order during the cell division process. In this study, DNA microarrays were used to analyze yeast cultures during alpha factor arrest, elutriation, and arrest of a cdc15 temperature-sensitive mutant. Cox17 expression was found to fluctuate with the cell cycle, with a maximum induction of 3.7 and a maximum repression of 2.4. The results for Cox17 along with those genes showing similar expression patterns can be seen below: |
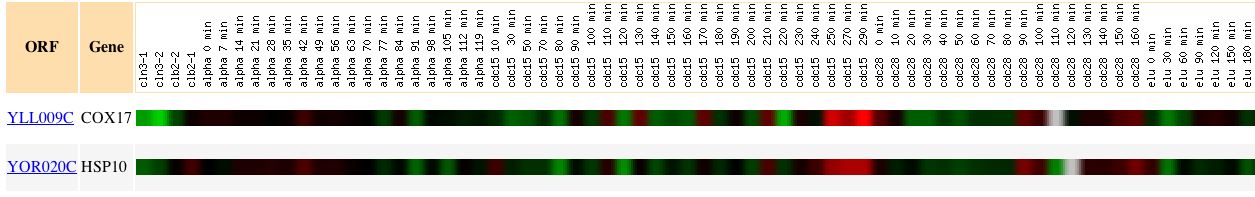 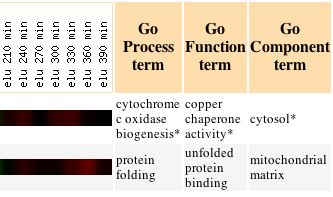 |
Figure 1: Expression pattern of Cox17 during the cell cycle. Image obtained from Expression Connection
With Cox17 being involved in cellular respiration, it is not surprising that its expression fluctuates with changes in the cell cycle. When a yeast cell is growing and dividing, it is likely that respiration rate will increase, thus placing a higher demand on the proteins involved in this process. It is particularly interesting to note that Cox17 is induced most at the end of the mitotic cycle (cdc15 290 minutes), perhaps when the need for energy is the greatest. It is also interesting to note that the only other gene found to have similar expression to Cox 17 is HSP 10, which is involved in protein folding in the mitochondrial matrix. Finding that Cox 17 exhibits a similar expression pattern to a mitochondrial gene during the cell cycle provides further evidence to support the likely association of Cox17 with the mitochondria as a copper chaperone. |
Expression during the diauxic shift:
In a study done by DeRisi et al. in 1997, expression profiles of genes with known metabolic functions were observed the diauxic shift, which is the change from anaerobic fermentation of glucose to aerobic respiration of ethanol. This “shift” occurs as glucose is being depleted and is known to be accompanied by major changes in gene expression, particularly in those genes involved in metabolic pathways. It is not surpising that Cox17 expression is induced with the shift from anaerobic to aerobic respiration, as the gene is directly involved with respiration. In fact, yeast that lack Cox17 are not capable of respiration (Heaton et al. 2000). The expression pattern of Cox17 during the diauxic shift can be seen below: |
 |
Figure 2: Expression pattern of Cox17 and similarly expressed genes during the diauxic shift. Image obtained from Expression Connection
It is also interesting to note that Cox17 shows a similar expression pattern to several other genes associated with the mitochondria (OXR1, YLR283W, YKL162C, SYM1, OCT1) and two proteins known to be involved in transport (PMP3, PEP12). The fact that proteins with similar functions and organelle associations are exhibiting similar expression patterns to Cox17 further affirms its role as a copper chaperone to the mitochondria. |
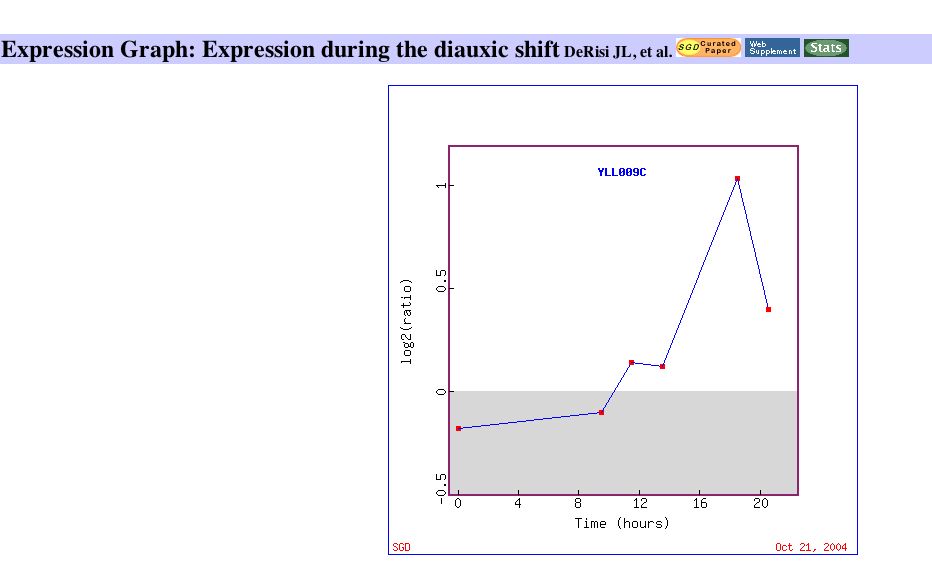
Since Cox17 is directly involved in yeast respiration, a more detailed view of this data is appropriate. An expression graph which plots the induction of Cox17 over time during the diauxic shift indicates that Cox 17 is induced most as glucose is depleted. The paper identified Cox17 to be part of a group of genes that is induced with a peak mRNA level around 18.5 hours. The expression graph can be viewed below: |
Figure 3: Expression of Cox 17 during the diauxic shift. Image obtained from Expression Connection.
It is clear from the expression data that Cox17 is part of a metabolic pathway that is induced by the diauxic shift, which is consistent with its role in respiration. |
Expression in response to environmental changes
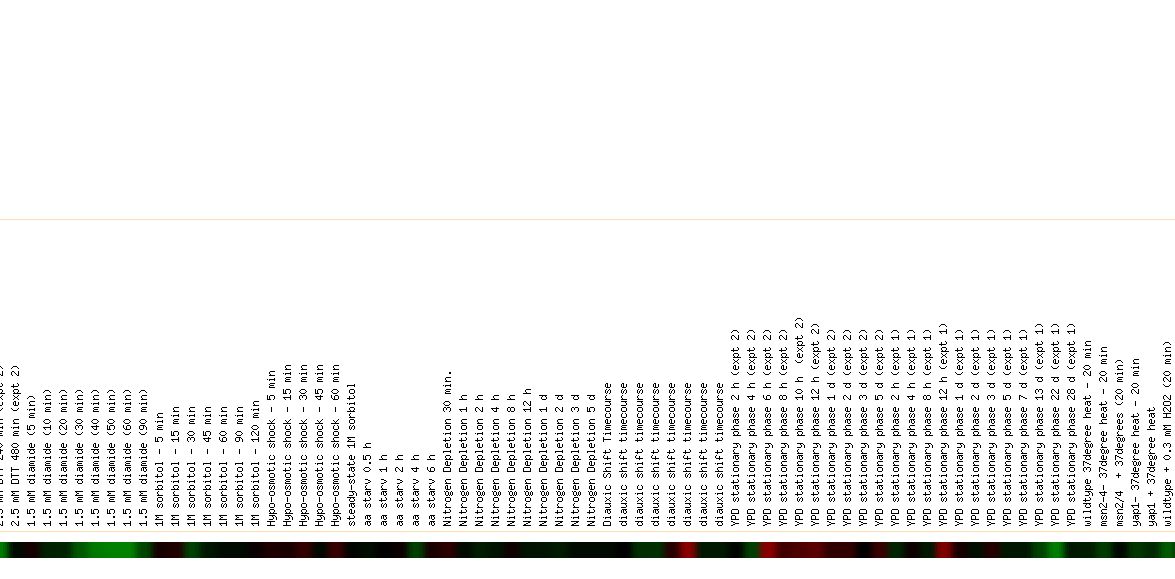
In an experiment conducted by Gasch et al. in 2000, the mRNA levels of various yeast genes were measured in response to a diverse panel of environmental stresses. The paper describes the global expression programs in response to a diverse set of stresses, including their specific features and a common response to all of the stressful conditions, termed the "environmental stress response" (ESR). The paper also explored the notion that several sets of coregulated genes share promoter elements, which point to the involvement of specific transcription factors in the regulation of those genes. Those genes induced during the environmental stress response included those involved in metabolite transport, vacuolar and mitochondrial functions, thus it is no surprise that Cox17 was so highly induced in this investigation. Many of the genes induced in the ESR have previously been proposed to offer cellular protection during stressful conditions, such as oxidative stress, heat shock, osmotic shock, and starvation. The data for Cox17 can be seen below: |
Figure 4: Expression pattern of Cox17 and similarly expressed genes in response to environmental changes. Image obtained from Expression Connection
The maximum induction of Cox17 (6.1 fold) occured while in the YPD stationary phase for 6 to 8 hours. In the absence of sufficient nutrients, cells will enter a stationary or G0 like phase. In the paper, it was noted that the genomic response to heat shock was strikingly similar to that triggered by stationary phase, including the induction of genes involved in respiration and alternative carbon source utilization (Gasch et al. 2000). With this information, it is not surprising that Cox17 was greatly induced during the stationary phase since it plays a critical role in respiration. |
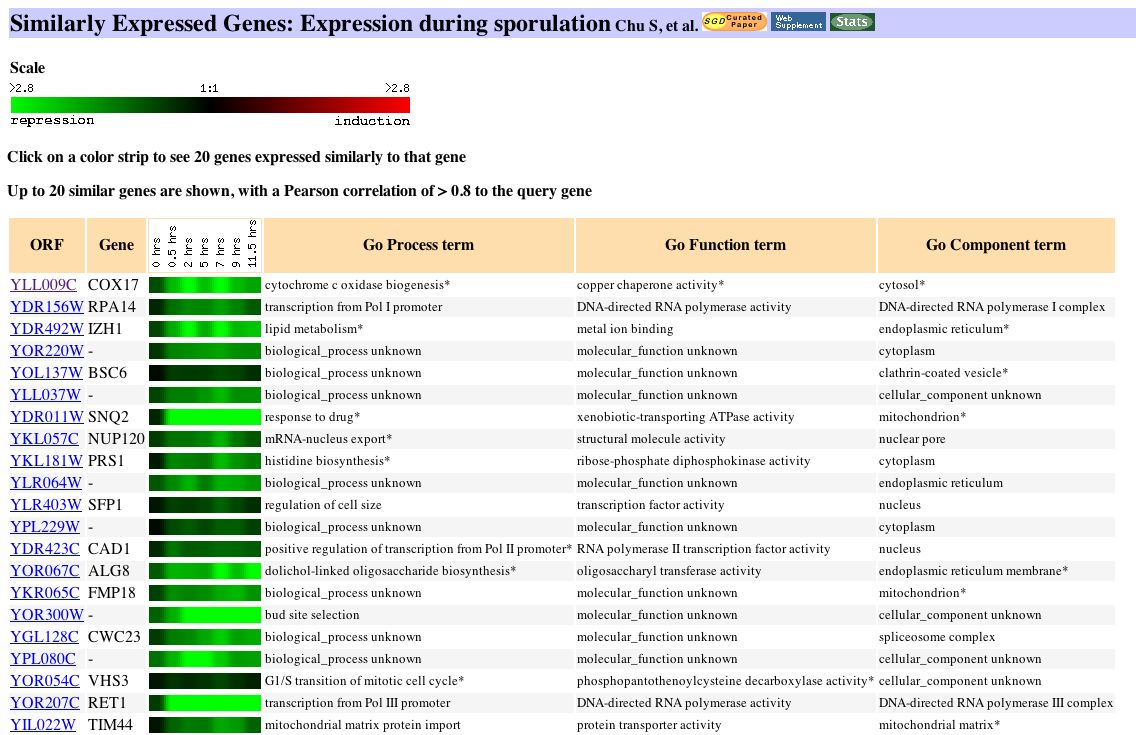
Expression during sporulation
The maximum repression of Cox 17 was seen in during sporulation (maximum repression = -2.8) in a study done by Chu et al. in 1998. Sporulation is the process by which diploid cells of budding yeast produce haploid cells. The expression pattern for Cox17 during sporulation can be seen below:
Figure 5: Expression pattern of Cox17 and similarly related genes during sporulation. Image obtained from Expression Connection.
Although it was not obvious from the paper why the Cox17 gene is repressed during a large portion of sporulation, it is possible that the repression is a result of nitrogen starvation, which induces sporulation. Nitrogen starvation is similar to many other stress responses, and from the expression pattern of Cox17 during environmental changes, it is clear that certain stress responses can significantly repress the Cox17 gene. |
|
Cox 17 Conclusions:
The expression patterns of Cox17 are supportive of the prediction that the protein is a copper charperone to the mitochondria and is an integral compenent of respiration in yeast. |
|
Analysis of YLL007C Expression Pattern:
Introductory Information:
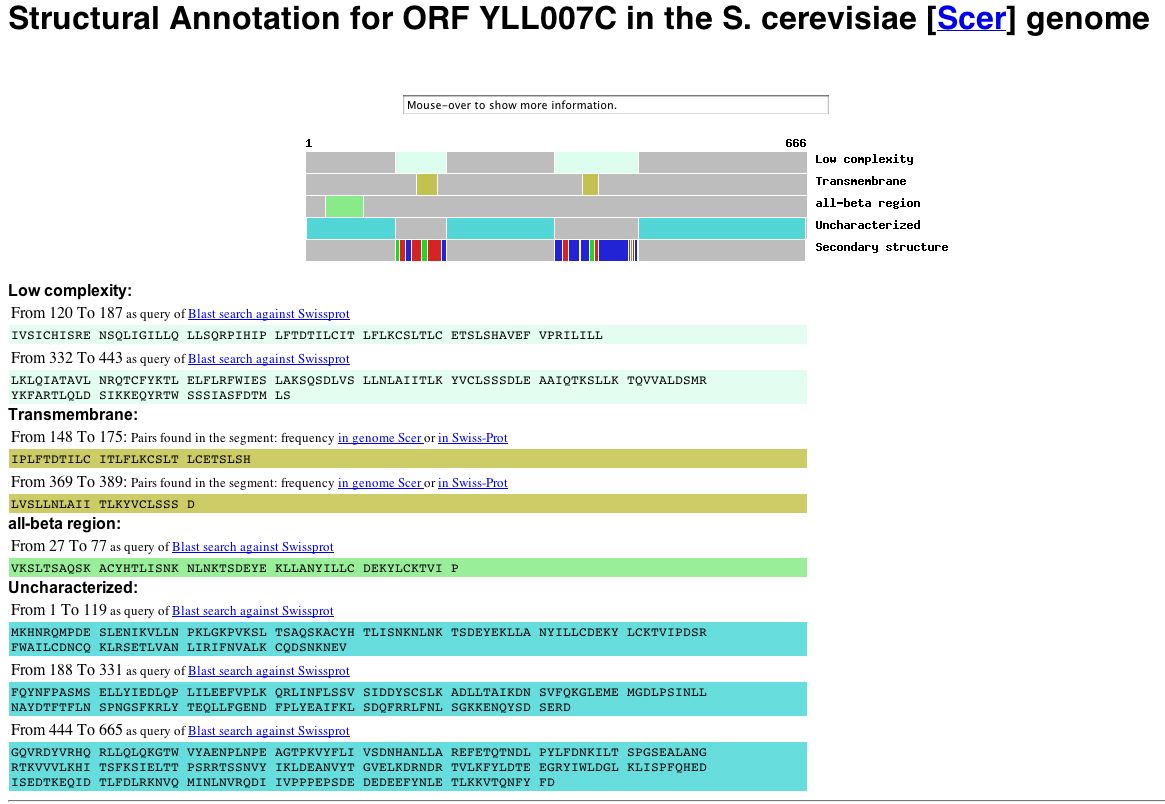
Data obtained from Microarray Global Viewer indicates that YLL007C is likely to have two transmembrane domains, as predicted in the previous assignment. The structural annotation for this gene can be seen below: |
Figure 6: Structural Annotation for the non-annotated yeast gene YLL007C. Image obtained from the Yale Gerstein Labs.
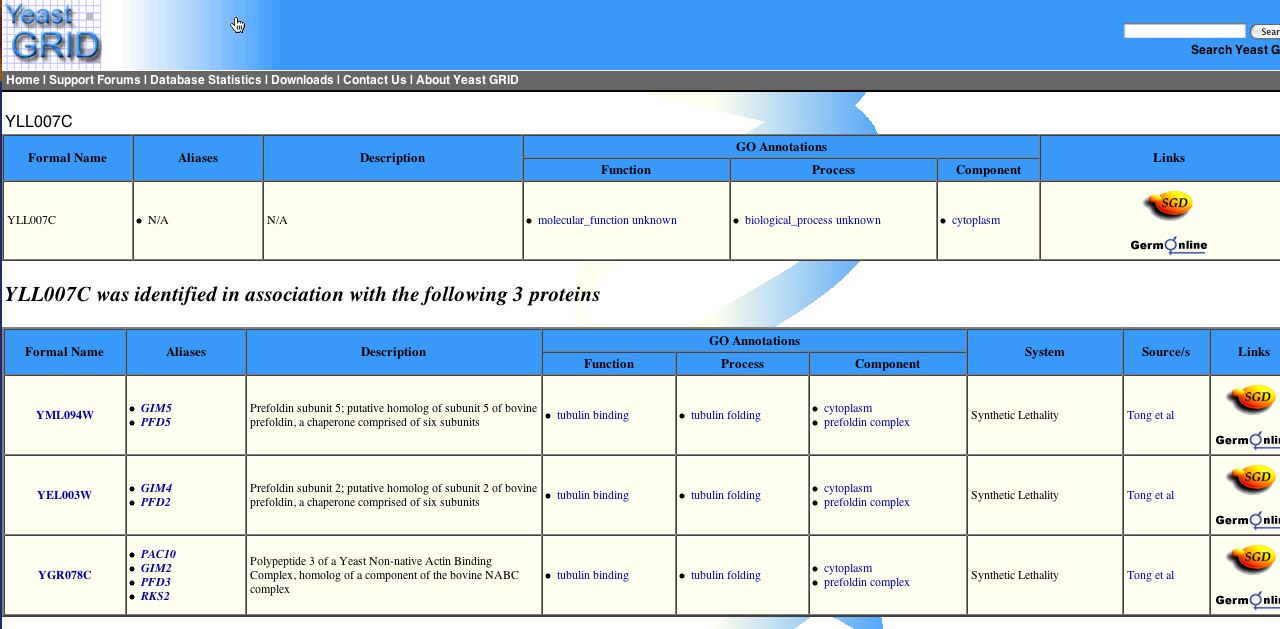
In addition to this data, I also found evidence using the Microarray Global Viewer that YLL007C is associated with three tubulin binding proteins as indicated in the figure below: |
| Figure 7: YLL007C is known to associate with three tubulin binding proteins. Image obtained from The Yeast Grid. |
YLL007C Expression Patterns:
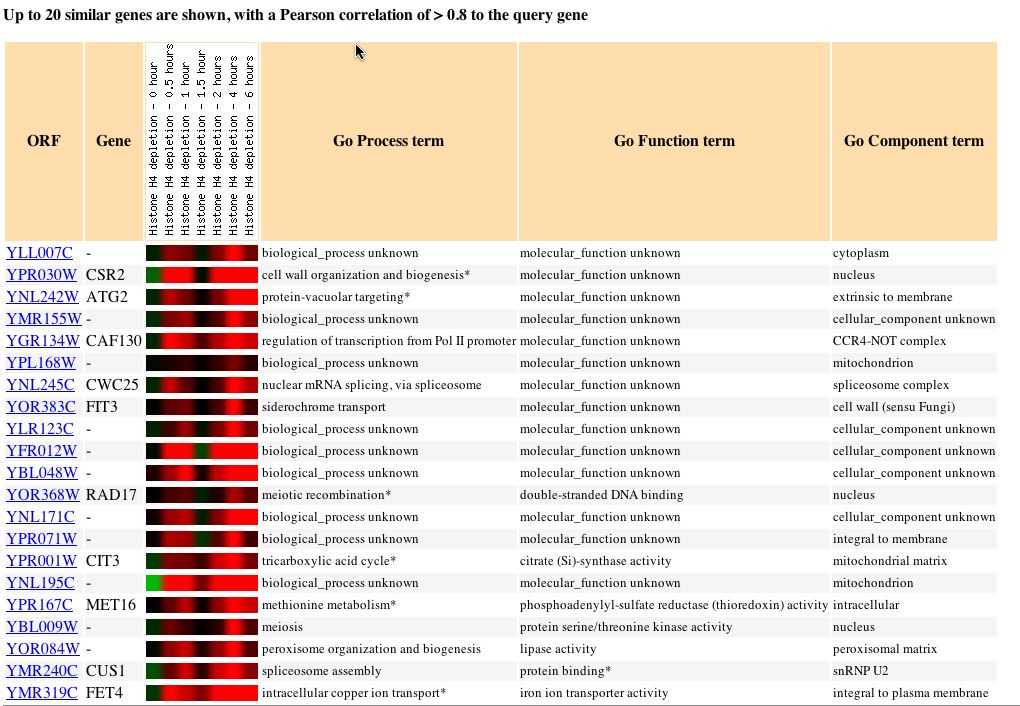
Using the Expression Connection Database, I determined under which conditions the YLL007C gene was most greatly induced. The YLL007C gene is most greatly induced under conditions of histone depletion. According to a study done by Wyrick et al. in 1999, reducing the nucleosome content by depleting histone H4 caused increased expression of 15% of genes and reduced expression of 10% of genes, but it had little effect on expression of the majority (75%) of yeast genes. The expression pattern for YLL007C along with similarly expressed genes under conditions of histone depletion can be seen below: |
| Figure 8: Expression pattern of YLL007C and other similarly expressed genes in response to histone depletion. Image obtained from Expression Connection. |
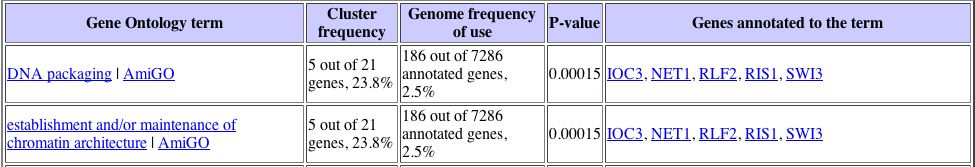
With this data, I proceeded to analyze the gene list with the Gene Ontology term finder. The Gene Ontology Term Finder compares the frequency of a particular biological process, function, or cellular component among the cluster of genes with the overall frequency of this term in the entire genome. The significance of these finding are indicated by P-values, with a smaller P-value corresponding to greater significance. The table below displays those shared Gene Ontology terms used to describe this set of genes that have a significant association. This data can be seen below: |
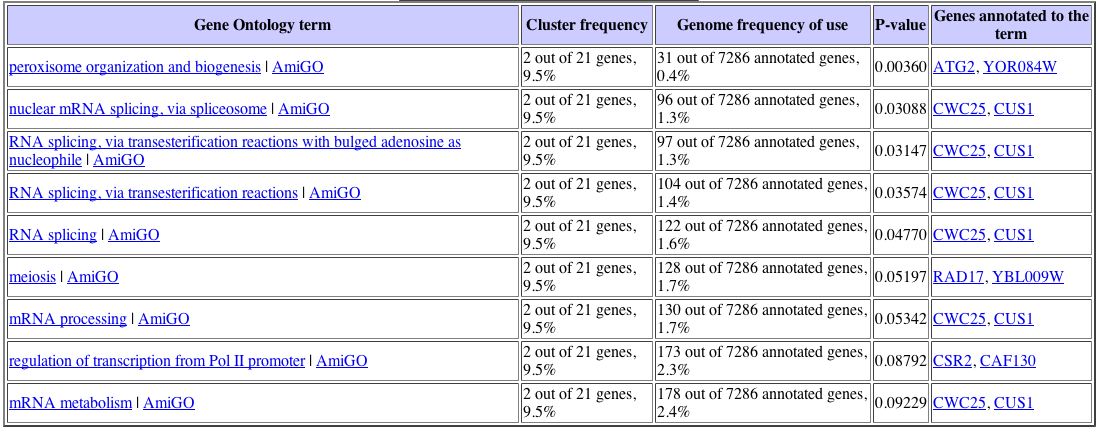
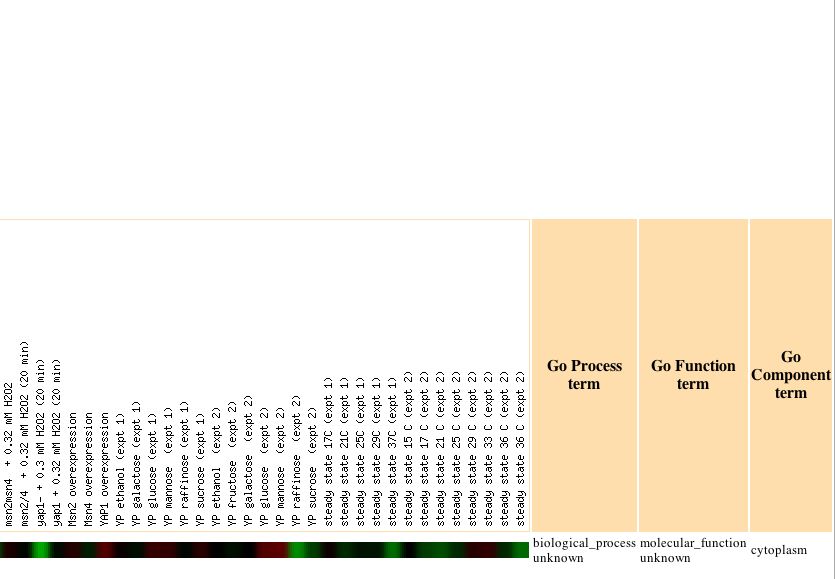
Unfortunately, only one Gene Ontology term proved to have a significant P value, which was peroxisome organization and biogenesis. Again turning back to the Expression Connection database, I determined under which conditions YLL007C was most greatly repressed. I found that YLL007C is repressed in response to particular environmental changes, with a maximum repression value of -2.9. The data for the expression pattern of YLL007C in response to environmental changes can be seen below:
|
| Figure 10: Pattern of Expression for YLL007C in response to environmental changes. Image obtained from Expression Connection. |
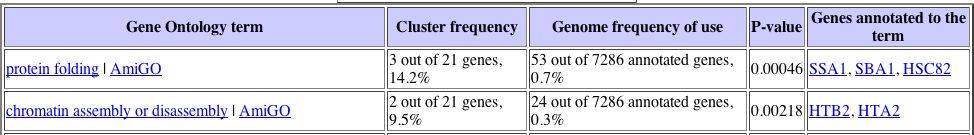
Unfortunately, there are no similarly expressed genes that are known under these particular conditions of environmental change, which makes it difficult to assign a cellular role to YLL007C gene based upon this information alone. There were several other circumstances, including during the diauxic shift, where YLL007C was the only gene found to express a particular expression pattern. I investigated the expression pattern of the YLL007C gene under several more conditions, not just those that were found to yield high maximum repression and induction values, and found some more conclusive results. The expression patterns for the YLL007C gene along with similarly expressed genes were analyzed using the Gene Ontology term finder to determine whether or not there were any significantly related Gene Ontology terms among those genes that were similarly expressed. Noteworthy results were found under conditions of glucose limitation and regulation by the PHO pathway. The Gene Ontology results can be seenbelow:
|
 |
|
It is interesting to note that both of these expression patterns returned a Gene Ontology term related to chromatin, which suggests that the YLL007C gene may have some nuclear role. |
YLL007C Conclusions:
In the previous assignment I hypothesized that YLL007C was a transmembrane protein involved in the regulation of the cell cycle. After analyzing expression data, some modifications must be made to this hypothesis. Keeping in mind the fact that mutations in the ORF of the YLL007C gene cause severe growth defects and taking into account the newly analyzed microarray expression data, I am able to suggest a more likely cellular role for this gene. Since YLL007C is likely a transmembrane protein and potentially associated with chromatin, it is possible that YLL007C is a nuclear membrane protein. It was indicated from Yeast Grid data that YLL007C was potentially associated with tubulin binding proteins. This association with tubulin could indicate that YLL007C is involved in nuclear migration during mitosis and mating. If this were the case, defects in YLL007C could result in a poor growth phenotype, and would thus be consistent with mutant data. |
References:
Beers J, et al. (1997) Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J Biol Chem 272(52):33191-6.
Campbell, et. al. (2003). Discovering Genomics, Proteomics, and Bioinformatics. Benjamin Cummings, pp. 107-117.
Chu S. et. al. (1998). The transcriptional program of sporulation in budding yeast. Science 282: 699-705.
DeRisi J.L. et. al. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-6.
Dunn B, Ferea T, Spellman P, Schwarz J, Terraciano J, Troyanovich J, Walker S, Greene J, Shaw K, DiDomenico B, Wang Q, Kaloper M, Metzner S, Chung E, Bondre C, Venteicher A, Botstein D, Brown P (2004) Genetic footprinting: A functional analysis of the S. cerevisiae genome
Gasch A.P. et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241-57.
Heaton D, et al. (2000) Mutational analysis of the mitochondrial copper metallochaperone Cox17. J Biol Chem 275(48):37582-7.
Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9(12):3273-97
Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA (1999). Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402(6760):418-21
|