This page was produced as an assignment for an undergraduate course at Davidson College.
My Favorite Yeast Expression:
OLE1 and YGL050W
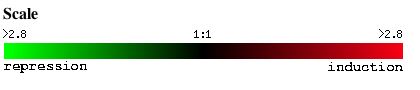
The following webpage elucidates expression patterns of annotated S. cerivisiae gene, ole1, and non-annotated gene, ygl050w, under various conditions. It draws information from microarray data obtained from Expression Connection, SGD which is based on the scale shown in Figure 1.

Figure 1. Scale used for all microarray data obtained from Expression Connection.
The Annotated Gene: OLE1
OLE1 Encodes stearoyl-CoA desaturase, an enzyme whose molecular function is to catalyze the following reaction (SGD):
Stearoyl-CoA + Reduced Acceptor + O2 ----> Oleolyl-CoA + Acceptor + 2 H20
It is a part of the lipid bilayer that surrounds the endoplasmic reticulum ( SGD). It is involved in the biological process of desaturation: it serves to form double bonds in the hydrophobic chains of saturated fatty acids by the removal of hydrogen (SGD). It is also involved in the distribution of mitochondria and mitochondrial genomes into daughter cells after mitosis or meiosis, mediated by interactions between mitochondria and the cytoskeleton (SGD).
Figure 2. List of yeast proteins that are functionally linked to stearoyl-CoA desaturase (modified from Expression Connection).
Figure 2 shows that stearoyl-CoA desaturase is linked to cytochrome-c isoform 1 (YJR048W) and cytochrome-c isoform 2 (YEL039C) whose deficiency results in cytochrome c deficiency (Function Junction). It is also linked COQ6 monooxygenase whose deficiency leads to a phenotype that is unable to produce ubiquinone and is hypersensitive to polyunsaturated fatty acid treatment (Function Junction). Stearoyl-CoA desaturase is known to transfer electrons from an oxygen molecule to cytochrome b5, an electron acceptor, it makes sense that it is associated with cytochrome c which is also an electron acceptor.
Figure 3. Expression graph of ole1 during sporulation (Expression Connection).
Figure 3 shows the expression of ole-1 during sporulation first increases slightly and then steadily decreases over time. In order to deal with unfavorable conditions, yeast cells undergo sporulation which involves meiosis and spore morphogenesis (Chu et al. 1998). Saccharomyces cerevisiae cells derepress expression of OLE1 encoding stearoyl-CoA desaturase under hypoxic conditions to allow more-efficient use of limited Oxygen (Nakagawa et al. 2001.). Thus, if stearoyl-CoA desaturase is responsible for increasing the degree of unsaturation in the cell membrane, it follows that during sporulation when the cell membrane is undergoing several changes, ole1 is not active since there is possibly inadequate energy for its function.
This hypothesis is supported by the microarray data shown in Figure 4 which shows maximum repression of ole1 (also called YGL055W). The second gene, pam1, whose expression is most similar to ole1, is connected to the bud neck which is most likely the small part of the cell membrane that connects the parent yeast to the forming bud temporarily. This, too falls into place, as budding, a process of asexual reproduction in yeast, will not under when sporulation is occuring. Cat5, a gene involved in ubiquinone metabolism, is also repressed. The expression of both pam1 and cat5 is repressed, although it is gradual over time, as indicated by the increasing intensity of the green in the microarray data for each of these in Figure 4.
Figure 4. Microarray data for expression of ole1 and similarly expressed genes during sporulation (Expression Connection).
The alpha-factor is produced by MAT-alpha cells, a strain of Saccharomyces cerivisiae cells (Son et al. 2004). The response to alpha-factor by cells of the opposite mating type, MAT-a, involves the formation of an outward projection called a shmoo (Vallier et al. 2002). Figure 5 includes microarray data that shows that ole1 expression is less repressed in the presence of alpha-factor over time for the first 50 minutes and then drops off with continued exposure. The increase can be accounted for the fact that shmoo formation occurs during mating, the predominant means of reproduction during favorable conditions such as availability of glucose, oxygen, etc. Once a shmoo has been formed, however, there is probably no need for stearoyl-CoA to be be expressed and thus ole1 expression drops off. Figure 6 shows shows similar expression patterns among some other genes that all appear to be repressed to more or less the same extent.
Figure 5. Expression graph of ole1 during exposure to alpha-factor over time (Expression Connection).
Figure 6. Microarray data for expression of ole1 and similarly expressed genes during exposure to alpha-factor over time (Expression Connection).
Thus, the microarray data, coupled with information obtained from the SGD, can help draw inferences about expression patterns of annotated genes in S. cerivisiae.
The Non-Annotated Gene: YGL050W
As determined from the previous webpage on My Favorite Yeast Genes, ygl050w may be a transmembrane protein. A search on Expression connection showed no known interactions of the gene product of ygl050w as seen in Figure 7 (Function Junction). GenomeNet, too, does not yield a known pathway for ygl050w (GenomeNet).
Figure 7. Interaction map of gene product of ygl050w (Expression Connection).
Figure 8 shows gene products that are functionally related to ygl050w. As several of these appear to be ribosomal proteins (YBL072C, YDL081C and YER102W) or translation initiation factors (YMR260C and YLR291C), it is likely that the gene product of ygl050w is somehow involved in translation.
Figure 8. List of genes that are functionally related to ygl050w.
The expression graph for expression of ygl050w during sporulation in Figure 9 shows that the ygl050w is repressed initially, then induced and repressed eventually. The microarray data obtained for ygl050w and similarly expressed genes during sporulation show the same pattern in Figure 10. If ygl050w were involved in translation, it is possibly participating in the production of proteins essential for spore formation or thereafter. Sporulation involves meiosis as genes are replicated and sorted into spores (Chu et al. 1998). It is therefore, possible that the ygl050w gene product is involved in translation of proteins involved in DNA replication.
Figure 9. Expression graph ygl050w during sporulation (Expression Connection).
Figure 10. Microarray data for expression of ygl050w and similarly expressed genes during sporulation (Expression Connection).
In the presence of alpha-factor, the expression of ygl050w increases rapidly and then drops off over time as can be seen from Figure 11. This implies that ygl050w is also involved in the mating response. If ygl050w is involved in translation, it might be involved in translation of proteins involved in shmoo formation during mating or perhaps, even mating itself. More inferences can be drawn from Figure 12 which shows microarray data for ygl050w. For instance, ylr107W, which shows very similar expression to ygl050w, is involved in RNA processing, a step in protein translation. However, other genes that show similar expression under the same conditions are also involved in proteolysis and peptidolysis (ygr232w and ygr 231c), antagonistic processes to protein production within the cell. Moreover, several of the genes that show similar expression in the presence of alpha factor have not yet been annotated, making it difficult to determine the likely identity and function of ygl050w.
Figure 11. Expression graph of ygl050w expression during exposure to alpha-factor over time (Expression Connection).
Figure 12. Microarray data for ygl050w and similarly expressed genes during exposure to alpha-factor over time (Expression Connection).
A Function Junction search for ygl050w homologs shows that ygl050w has no known homologs in yeast or worm, and does not appear in any shared or unshared clusters.
Conclusion
While it is possible to explain the role of stearoyl-CoA desaturase in the life of Saccharomyces cerivisiae, ygl050w appears cryptic. Its relationship to other yeast proteins hints that it might be a translation factor. It is unclear exactly what role it plays in translation. It might be responsible for translation proteins in the cell membrane as it is induced both during sporulation and during mating, two distinct processes that occur under an opposing set of conditions. On the other hand, given that protein metabolism genes appear to be expressed under the same conditions to the same degree, it is difficult to make a confident conclusion about the role of the gene product of ygl050w. Moreover, the fact that there are currently no known homologs and that ygl050w does not show up in any clusters further clouds the possible role and identity of ygl050w.
References
Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P.O., and I. Herskowitz. 1998. "The Transcriptional Program of Sporulation in Budding Yeast." Science. 282: 699-705.
Expression Connection. 2004. <http://www.genome.jp/> Accessed 2004 Oct 21.
Function Junction. 2004. <http://db.yeastgenome.org/cgi-bin/SGD/functionJunction> Accessed 2004 Oct 20.
Nakagawa, Y., Sugioka, S., Kaneko, Y., and S. Harashima. 2001. "O2R, a novel regulatory element mediating Rox1p-independent O(2) and unsaturated fatty acid repression of OLE1 in Saccharomyces cerevisiae." Journal of Bacteriology. 183(2):745-51.
SGD. 2004. <http://www.yeastgenome.org/> Accessed 2004 Oct 20.
Son, C. D., Sargsyan, H., Naider, F., and M. Jeffrey. 2004. " Identification of Ligand Binding Regions of the Saccharomyces cerevisiae R-Factor Pheromone Receptor by Photoaffinity Cross-Linking." Biochemistry. 43:13193-13203.
Vallier LG, Segall JE, and M. Snyder. 2002. “The alpha-factor receptor C-terminus is important for mating projection formation and orientation in Saccharomyces cerevisiae.” Cell Motil Cytoskeleton. Dec;53(4):251-66.