
The Proteins of ADY2 and YCR007C
This web page was produced as an assignment for an undergraduate course at Davidson College.
This web page was designed to discover as much as possible about the proteins encoded for by ADY2 and YCR007C.
ADY2- An Annotated Gene
Molecular Function: ADY2 codes for the transmigrate Ady2p protein which directs the active transport of several macromolecules, small molecules, and ions across, between or within cells. Biological Process: This gene has several biological processes. First, it plays a major role in DNA metabolism, involved in the accumulation of DYads, and therefore important in meiosis. ADY2 is also involved in acetate transport, and is essential in acetate permease activity. A third biological process that this gene contributes to is nitrogen utilization and ammonia production. Ammonia is important as a starvation signal between yeast colonies, as discussed by Palkova, et al. (2002, <source>) Cellular Component: Ady2 is a transmembrane protein and is also found in the mitochondrion. 3D Structure: Unfortunately, the 3D structure of Ady2p is not known. |

Fig. 1: Protein sequence of Ady2p.
The scientists that established the TRIPLES database inserted transposons to disrupt genes and to see what the effects of mutations have on yeast phenotypes.
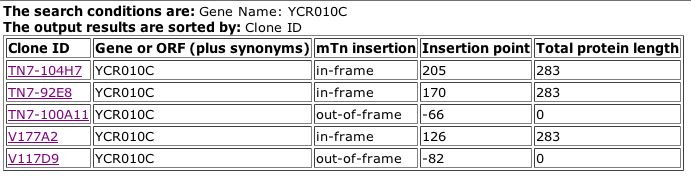
Fig. 2: Results from searching the TRIPLES database for ADY2, aka YCR010C.
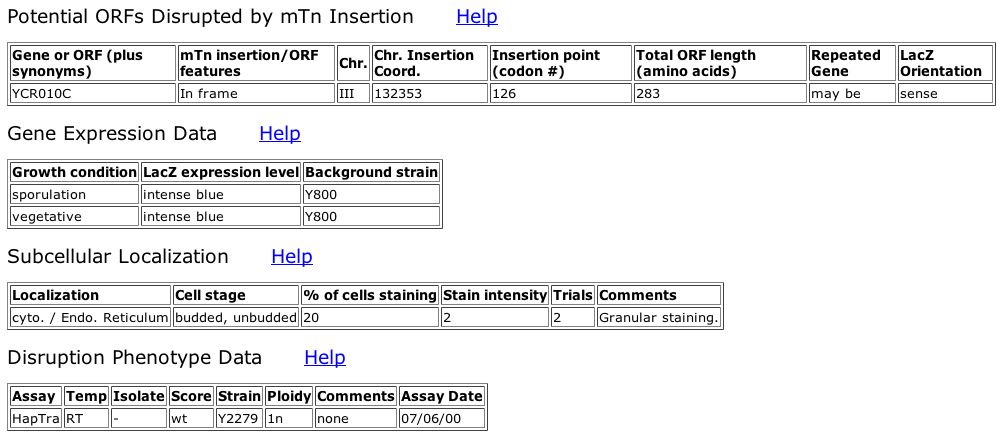
Fig. 3: Information obtained from the 4th clone ID, V177A2.
| From the pictures above, we can see that Andy is expressed during both sporulation and vegetative growth states. This shows that it is an important protein throughout the cell's entire life cycle. We can also see that there is no significant phenotypic difference between the wildtype protein and this particular mutation, meaning that this mutation either does not affect the function of the protein, or there is redundancy within the proteome, with other proteins that are able to perform the same function. |
Protein Interactions:
Unfortunately, searching both DIP (Database of Interacting Proteins) and PathCalling, no protein interactions were found for ADY2p.

Fig. 4: Pathcalling search results showing no protein interactions.
Experiments:
There are several experiments that I would like to see done for Ady2p. First, no information was found on the Y2H database. I think it would be beneficial to use the yeast two-hybrid method, with Ady2p as the bait protein, to see what other proteins interact with it. I predict we would find other proteins involved in ion transport and DNA metabolism would interact with Ady2p, and we could gain insight into the the exact mechanisms of these processes in yeast. A second experiment that I would perform is to create a knockout strain of this gene and study what effects that has on the organism. Studying the resulting phenotype would provide us with further insight into the role Ady2p plays in the biological processes it is associated with. We also learned from the SGD that an ADY2 mutant is viable, but perhaps if it is tested in different media, it would be affected differently. It would also be interesting to see a protein microarray, through the use of antibodies, to detect the amount of Ady2p in yeast populations grown in different media. Specifically, it would be beneficial to study populations grown in different nitrogen and ammonia medias, since ADY2 is known to regulate nitrogen utilization and ammonia production. Finally, the structure of this protein needs to be determined. Using the protein sequence, we can predict the 3D structure of this protein, and in this way visualize any possible interactions it has with other molecules. |
Databases Searched to no Avail:
Enzymes and Metabolic Pathways Database
Benno Experiment Results
YCR007C: An Unannotated Gene
YCR007C is a suspected gene whose biological process, molecular function, and cellular component are unknown. In a previous assignment I hypothesized that since it belongs to the DUP240 gene family, it is likely to have similar functions. The two known genes in this family have as their biological function protein binding, the process they accomplish is vesicle organization, and their cellular components are the golgi apparatus, the endoplasmic reticulum, and the plasma membrane. 3D Structure: Unfortunately, the 3D structure of the YCR007C protein is not known. |

Fig. 5: Protein sequence of unannotated gene YCR007C.
Using the TRIPLES database once again, the following information was found:
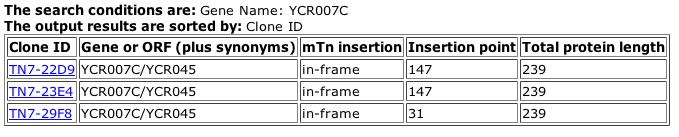
Fig. 6: Results from searching the TRIPLES database for YCR007C.
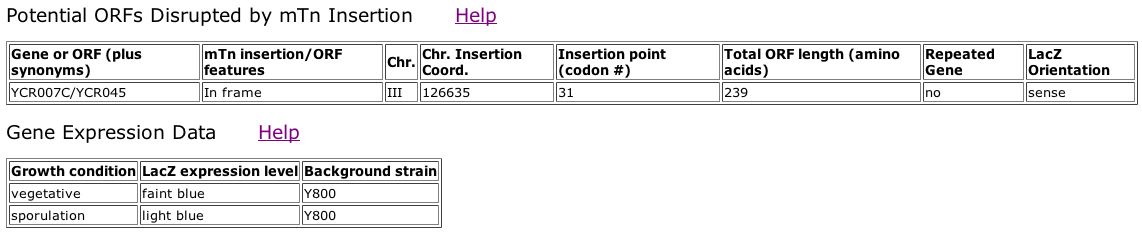
Fig. 7: Information obtained from the third clone ID, TN7-29F8.
| From the images above, we can see that YCR007C is slightly expressed during sporulation, and barely expressed during the vegetative growth state. The TRIPLES database provided no information about the disruption of the phenotype in these YCR007C mutants. |
Protein Interactions:
The following images were obtained from the DIP Database:
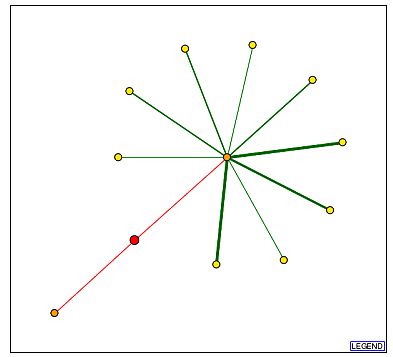
Fig 8: Graph of protein interactions for YCR007C and the legend.
| The YCR007C protein seems to have interactions with 2 main proteins. MEC3 is the protein with which it interacts that leads to the large number of secondary interactions. MEC3 is a DNA damage checkpoint protein whose cellular component is in the nucleus. (SGD) Of the proteins that interact secondarily with YCR007C, 5 are located in the nucleus, all of which are involved in one of the following processes: transcription, splicing, gene silencing and DNA damage checkpoints. ULP1p, the other primary node interacting with YCR007C in the figure above, is a nuclear membrane protein involved in the transition between the G2 phase to the M phase of the cell cycle. (SGD) If we use the guilt by association model for these protein interactions, we could assume that the protein encoded for by YCR007C has its cellular component in the nucleus and has a biological process involved in the mitotic cell cycle. This conclusion contradicts that arrived at in the two previous assignments where predictions were made based solely on gene expression information. |
These thoughts seem to be confirmed by the MIPS database:
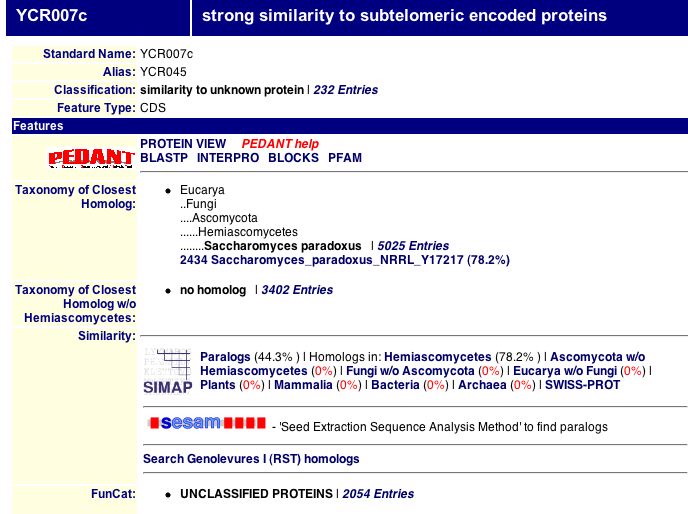
Fig. 9: Information on the MIPS database for YCR007C.
Experiments:
Since I found no information for this protein on the Y2H database, this would be the first experiment I would perform on this gene as well. This would confirm the interactions between YCR007C and its two primary nodes, which are as of yet unverified, and would also identify any other proteins that it interacts with. The structure of this protein also needs to be determined. By using the protein sequence, researchers could make predictions of the 3D structure of the protein, and this would help in predicting the function of the protein. These theories could then be tested experimentally, and we would know if this protein is an important link in the mitotic cell cycle. Though I am not sure of the experimental design, we need to determine the cellular component of this protein. Knowing where a protein functions is very important in determining its role in the cell. Creating a knockout strain of this gene would also provide us with information on the function of this protein in the cell. Previous studies found that the cell is viable with the deletion of this gene, but it would be interesting to determine whether it is able to go through mitosis. If the prediction above is correct and YCR007C has a role in mitosis, these cells would not be able to divide mitotically, even if they were able to mate successfully. |
Databases Searched to no Avail:
Enzymes and Metabolic Pathways Database
Benno Experiment Results
Davidson College Biology Department
Resources:
[DIP] Database of Interacting Proteins. 2004 Nov 18. http://dip.doe-mbi.ucla.edu/dip/Search.cgi?SM=3
[MIPS] Munich Information Center for Protein Sequences. 2004 Nov 18. http://mips.gsf.de/genre/proj/yeast/index.jsp.
[SGD] Saccharomyces Genome Database. 2004 Nov 18. SGS1/YMR190C. http://db.yeastgenome.org.
[TRIPLES] TRansposon-Insertion Phenotypes, Localization, and Expression in Saccharomyces. 2004 Nov 18. http://ygac.med.yale.edu/triples/basic_search.asp.
Campbell AM, Heyer LJ. 2003. Discovering Genomics, Proteomics, and Bioinformatics. Benjamin Cummings: San Francisco.