*This page was created as an undergraduate assignment for a Genomics course at Davidson College*
YNK1 and YKL069W
My Favorite Yeast Genes
Introduction
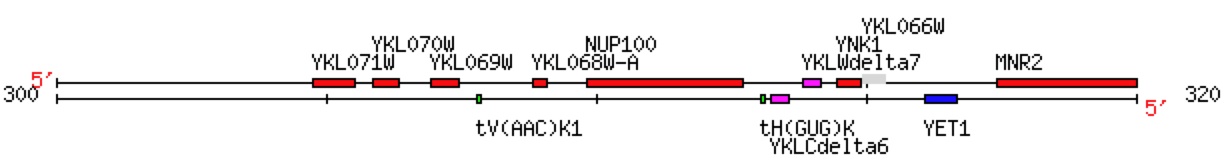
YNK1 is an annotated gene that encodes Nucleoside diphosphate kinase, a protein whose function is to phosphorylate nucleosides into their triphosphate versions (Jong and Ma, 1991). YNK1 and YKL069W are both located on chromosome 11 of the Saccharomyces cerevisiae genome (figure 1). YKL069W is a non-annotated gene nearby YNK1 that has no known molecular function, or cellular component, or related biological processes.

Fig. 1. A map of a section of Chromosome 11 in which YNK1 and YKL069W are both located on the Watson strand of the DNA. Image from Saccharomyces Genomic Database (http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000001550).
YNK1 (my favorite annotated yeast gene)
Biological Process
YNK1 encodes nucleoside diphosphate kinase an enzyme responsible for the phosphorylation of nucleoside diphospates to triphosphates that can then be used in DNA biosynthesis (Jong and Ma, 1991). This is a very important functions as DNA synthesis could not occur with the proper balance between nucleoside diphosphates (NDP) and nucleoside triphosphates (NTP) (Amutha and Pain, 2003).
Biological Function
The YNK1 protein removes the gamma phosphate from one NTP and places it on a NDP. The enzyme accomplishes this by removing the phosphate from the NTP and phosporylating itself and then transfers this phosphate group onto the NDP. The YNK1 protein is constantly performing this action in order to make a uniform set of NDP's and NTP's (Amutha and Pain, 2003). YNK1 proteins have also been found to have a phosphotransferase activity, meaning they can phosphorylate other proteins, thus having potential effects on other cellular processes (Amutha and Pain, 2003).
Cellular component
The YNK1 protein is found in the cytoplasm, with small amounts being located in the mitochondria (Asmutha and Pain, 2003). It is believed that the mitochondrial recognition tag is buried within the tighyly folded protein, causing it to be located in the cytoplasm (Asmutha and Pain, 2003).Related Genetic Disorders
There are no known genetic disorders arising from a mutation in YNK1. There has been a study in which the gene was deleted and it was shown that the deletion was neither lethal, nor had an impact on the phenotype of the Yeast (Giaever et al. 2002 and Fukuchi et. al 1993). This is curious because there are no proteins found to be highly similar when the YNK1 sequence is used in a protein blast query (figure 2). This also proves that the YNK1 protein is not required to be viable.
Fig. 2. The results from a BLASTp, in which YNK1 sequence was used as the query sequence, showing the proteins that are most like the YNK1 protein product. Image found using BLAST at this wed address, http://www.ncbi.nlm.nih.gov/BLAST/
Other Interesting Info about YNK1
YNK1 is encoded on Chromosome 11 and is 462 base pairs long. The Protein encoded by YNK1 is 153 amino acids in length. To view both sequence click here.
Using the PREDATOR protein predictor, one can predict secondary structure of the YNK1 protein.
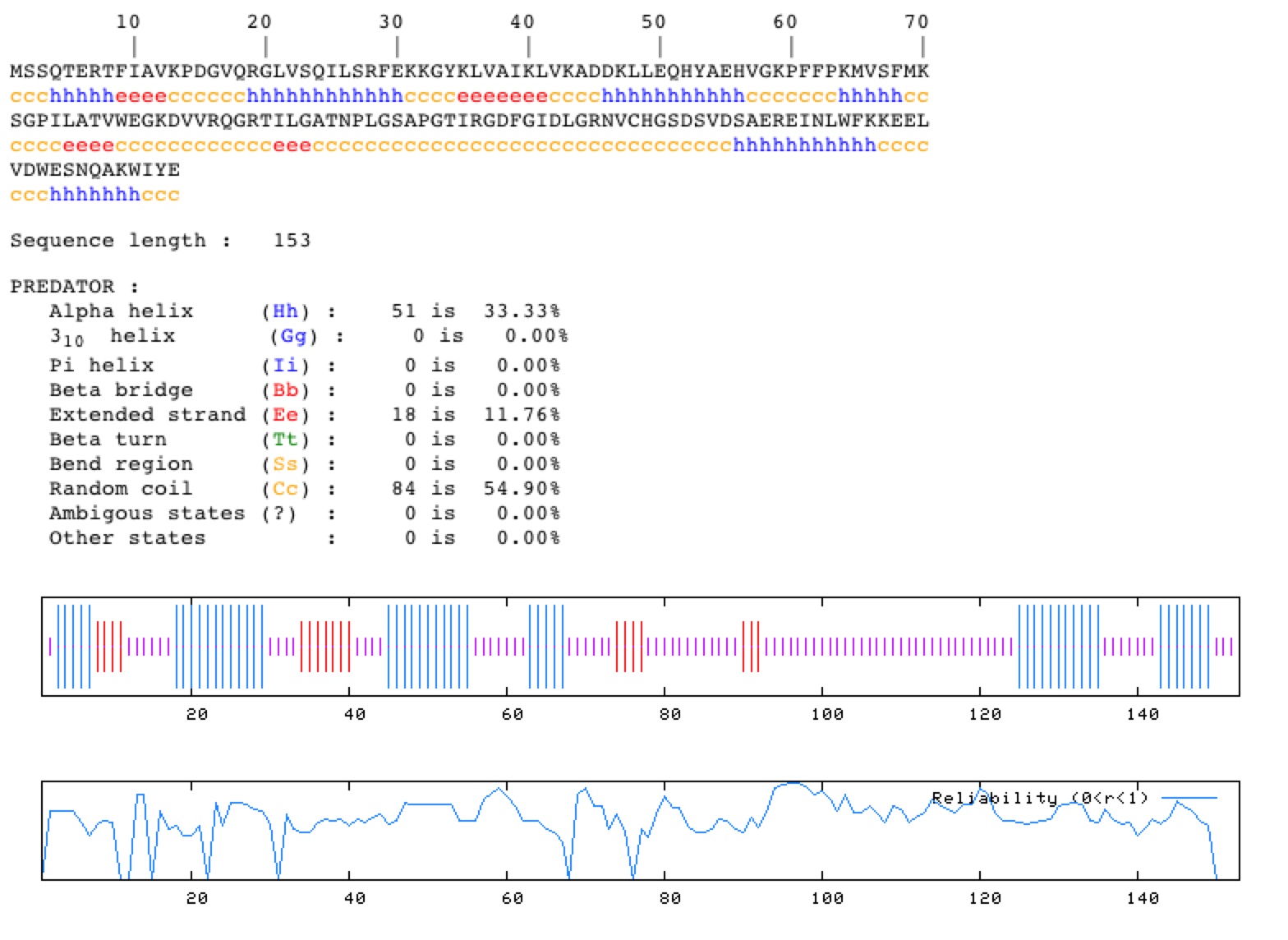
Fig. 3. This is a prediction made in silico by the PREDATOR prediction software, showing the predicted secondary structure of the YNK1 protein. It has each amino acid listed and the conformation that each of these is predicted to take. (http://npsa-pbil.ibcp.fr/) Permission Pending.
Since YNK1 protein is found mainly in the Cytoplasm, one would not expect to find any transmembrane domains and as you can see in this Kyte-Doolittle hydropagthy plot, no transmembrane domains would be predicted due to the hydropathy score never reaching above 1.7 (red line) (Figure 4).
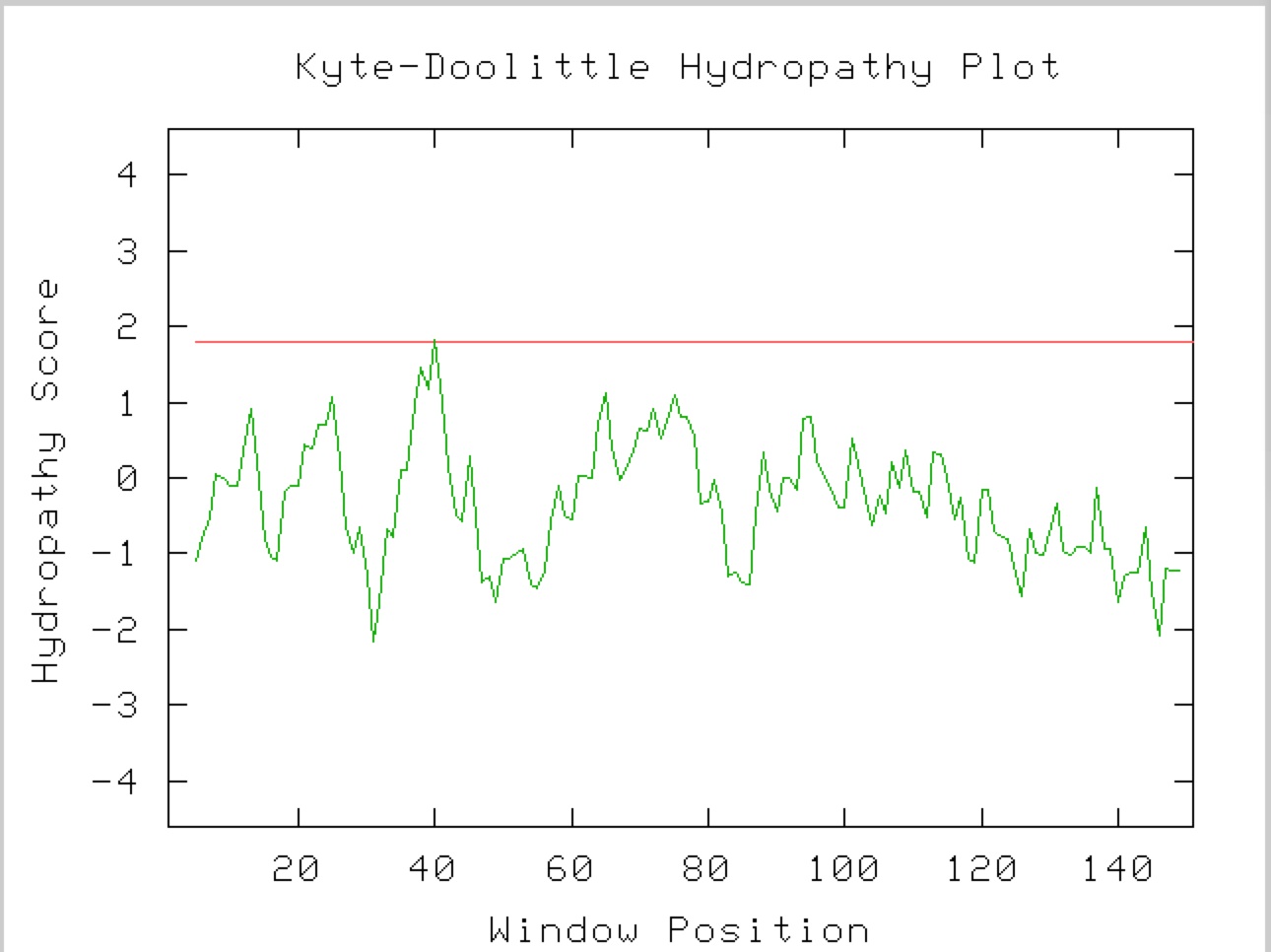
Fig. 4. This is a Kyte-Doolittle Hydropathy plot which predicts transmembrane domains for YNK1. Picture taken from Kyte-Doolittle, with the YNK1 as the query sequence.
An image for the Saccharomyces cerevisiae YNK1 protein could not be found at the Protein Data Bank, but the human version, which has a high similarity did contain a drawing (Figure 5)
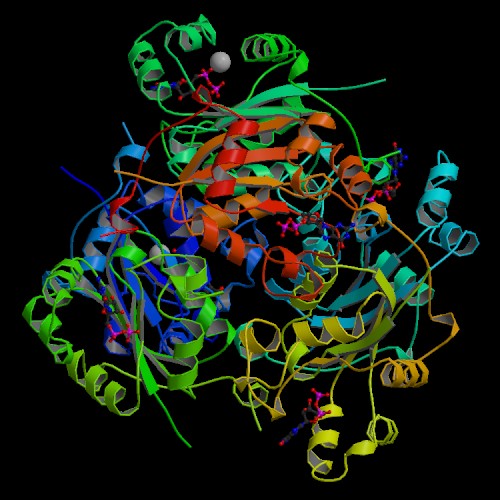
Fig. 5. This is an image taken from the PDB of the human version of Nucleoside Diphosphate Kinase as found at http://www.rcsb.org/pdb/.
The YNK1 protein is also associated with many other proteins, showing that it has the potential to have many functions. Not suprising because it is thought to be able to phosphorylate other proteins as well as nucleoside diphosphates (Figure 6).
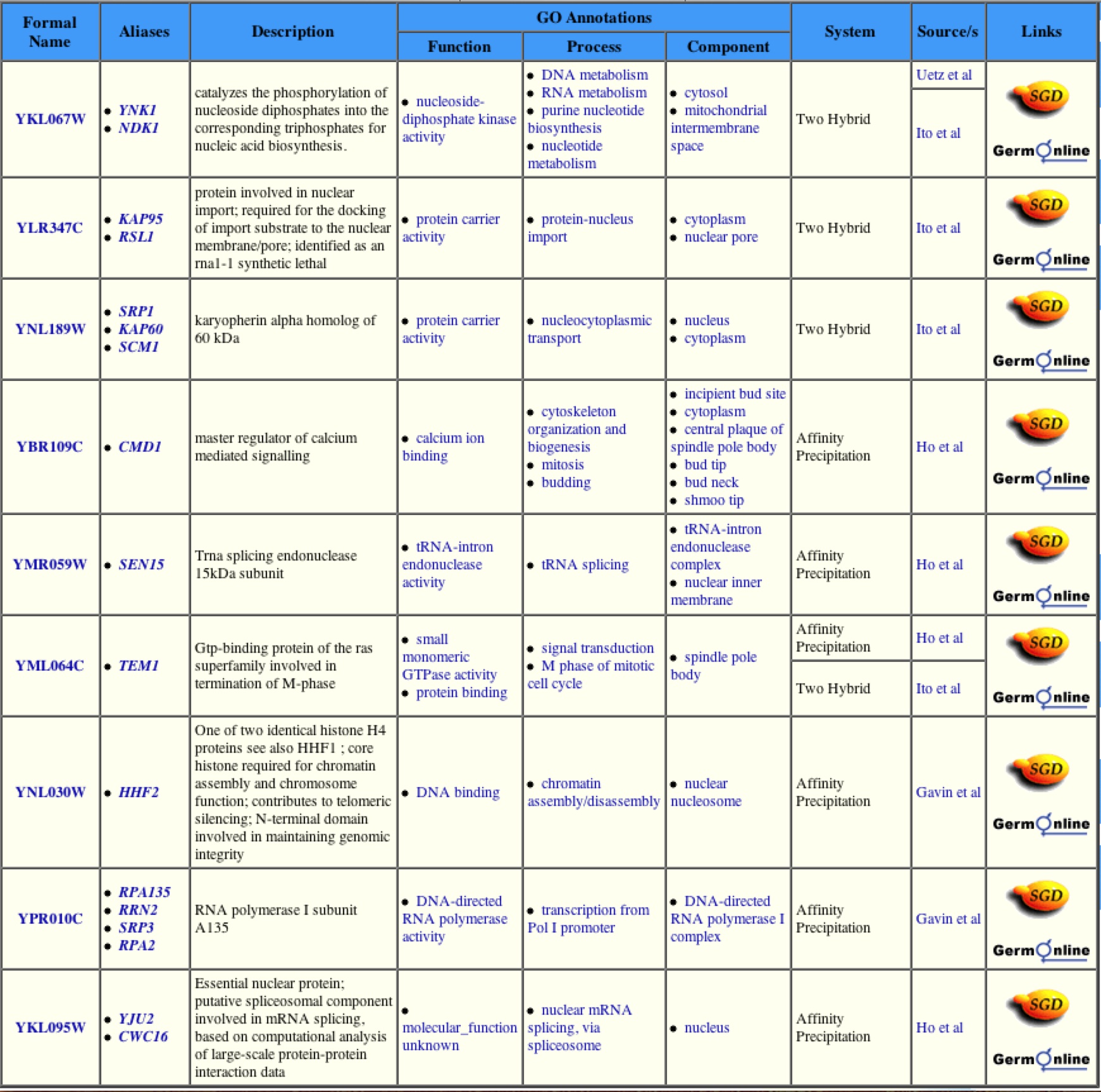
Fig. 6. This Table shows the various other proteins with which the YNK1 protein interacts. Due to the large number of interacting proteins, one can assume that the YNK1 protein may have many different functions. This image was taken from yeast grid.
YKL069W (my favorite non-annotated gene)
YKL069W can be located on Chromosome 11 in S. cerevisiae. It is located upstream of YNK1 and little is known about this gene (Figure 1). The biological process, biological function, and cellular component for this gene are all unknown. The DNA and protein sequences however are known. From these sequences I will try to hypothesize a function for the protein this gene would produce.
I will first use a Kyte-Doolittle Hydropathy Plot to determine if the protein has any transmembrane domains (Figure 7). Based on figure 7 there do not appear to be any transmembrane domains in the YKL069W protein.
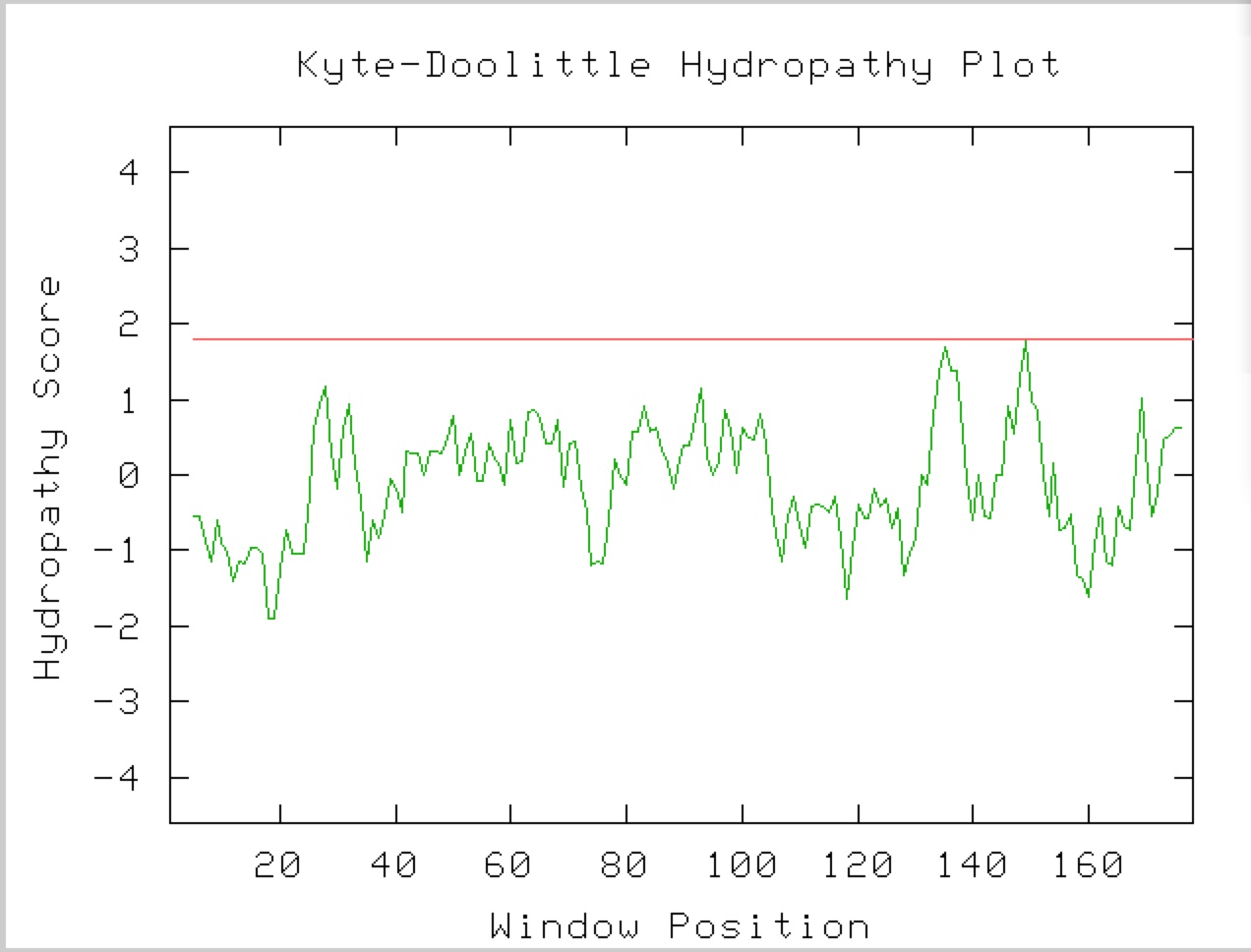
Fig. 7. This is a Kyte-Doolittle hydropathy plot of the YKL069W protein. This figure was taken from the Kyte-Doolittle with the YKL069W as the query sequence.
Next I will blast the sequence to determine if the protein sequence is similar to that of any known protein (Figure 8). The YKL069 protein appears to be related to several other proteins with no known fucntion, however if you look further down the list you find a few that say GAF domain containing proteins.
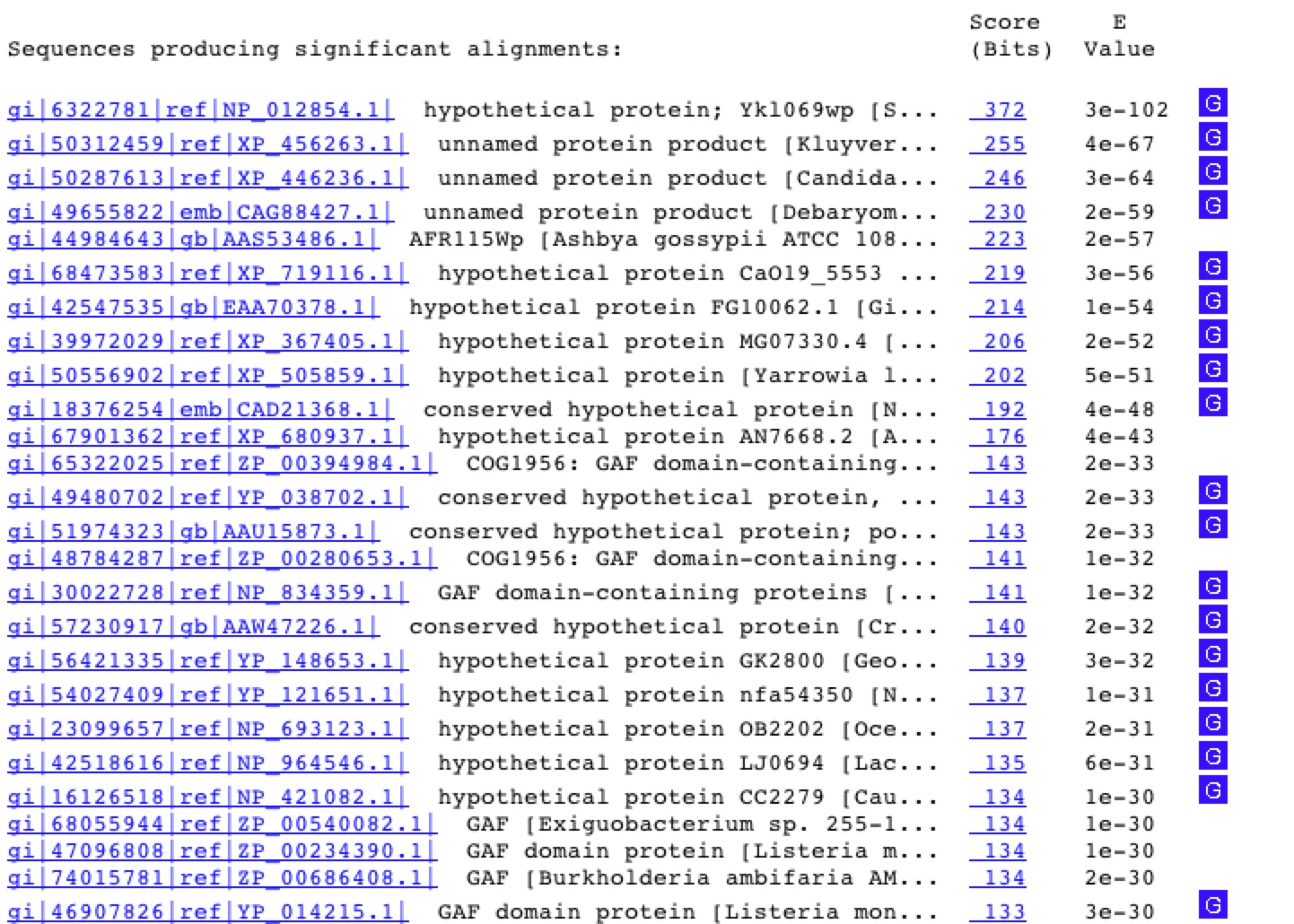
Fig. 8. These are the BLASTp resultss when the YKL069W protein sequence was used as the query. http://www.ncbi.nlm.nih.gov/BLAST/
GAF Domains
GAF domains are 110 amino acids sequences that are often found in cyclic nucleotide phosphodiesterases (Zoraghia et. al 2004, Martinez et. al 2002). These domains often lead to the binding of a cyclic nucleotide (Figure 9). These GAF domains are found in 1400 proteins predicted by a non-redundant database (http://smart.embl-heidelberg.de), leading one to believe that the GAF domain may have other functions that have not be previously charactertised (Zoraghia et. al 2004).
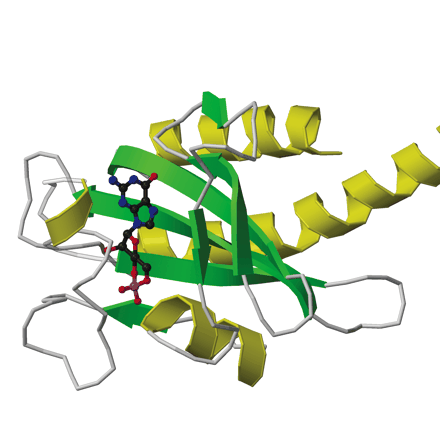
Fig. 9. This is an example of a GAF binding domain, that is bound to a cyclic nucleoside. This image was taken from the paper GAF Domains: Two-billion-year-old molecular switches that bind cyclic nucleotides. Permission Pending.
GAF domains have however been shown to bind many other molecules. Therefore it is not appropriate to assume that the YKL069W protein binds strictly to cyclic nucleotides. This does suggest that the protein produced by YKL069W does produce an enzyme.
PREDATOR in silico secondary struture of the YKL069W protein reveals that the protein contains a vast amount of random coiling as well as 26.11% Alpha helices (Figure 10).
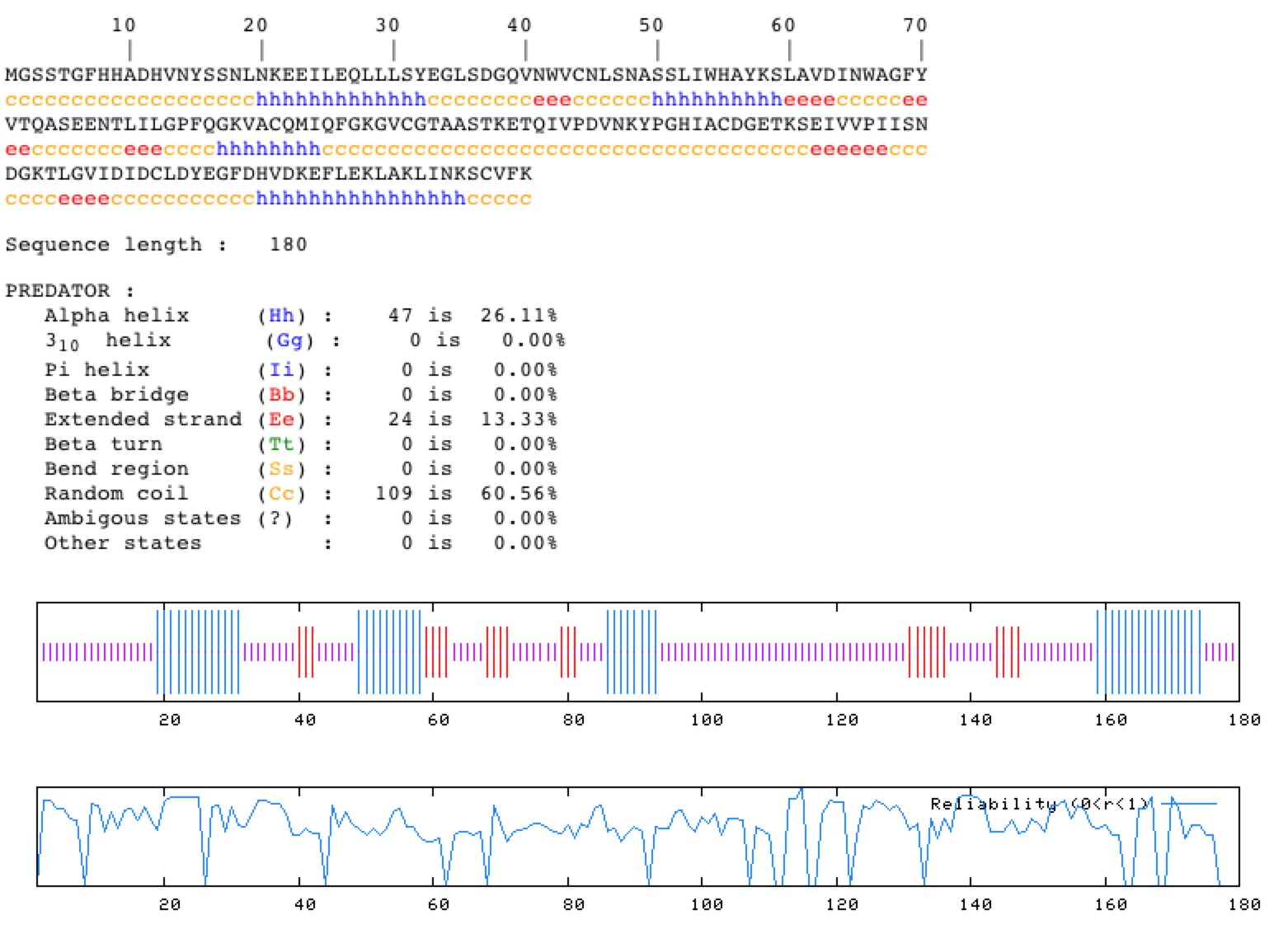
Fig. 10. The PREDATOR in silico secondary structure of the YKL069W protein. Image taken from (http://npsa-pbil.ibcp.fr/) Permission Pending.
Related Genetic Disorders
There is no information known about the function of this protein so no genetic disorders are directly attributed to the YKL069W protein. It is known that the deletion of the YKL069W protein is not detremental to the growth or viability of the S. cerevisiae (Giaever et al. 2002).
Predictions
The YKL069 protein in probably not an integral membrane protein due to the Kyte-Doolittle Hydropathy plot results. The YK069W protein does contain a GAF domain suggesting that its function could be in the creation of phosphodiesterase bonds or could be required in a signalling pathway to bind to cyclic nucleotides. The protein is most likely an enzyme found in the cytoplasm and performs its duties in this compartment.
References:
Amutha B, Pain D. 2003. Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. Biochemical Journal 370:805-815.
Dolinski, K. et. al. 2003. Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed 2005 5 Oct.
Fukuchi T, Nikawa J, Kimura N, Watanabe K. 1993 July. Isolation, overexpression and disruption of a Saccharomyces cerevisiae YNK gene encoding nucleoside diphosphate kinase. Gene 129(1):141-6.
Giaver et. al. 2002 July. Functional profiling of the Saccharomyces cerevisiae genome. Nature 18: 387-391.
Jong AY, Ma JJ. 1991 December. Saccharomyces cerevisiae nucleoside-diphosphate kinase: purification, characterization, and substrate specificity. Archives Biochemistry Biophysics 291(2):241-6.
Martinez SE, Beavo JA, Hol WGJ. 2002 September. GAF Domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Molecular Interventions 2(5): 317-323.
Mount Sinai Hospital. 2002. The GRID database. <http://biodata.mshri.on.ca/grid/servlet/Index> Acccessed 2005 5 Oct.
NCBI. 2003. National Center for Biotechnology Information. <http://www.ncbi.nih.gov/> Accessed 2005 5 Oct.
[PREDATOR] <http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html>. Accessed 2005 5 Oct.
Zoraghi R, Corbin JD, Francis SH. 2004. Properties and Functions of GAF Domains in Cyclic Nucleotide Phosphodiesterases and Other Proteins. Molecular Pharmacology 65:267-278.
Any questions or concerns regarding this page should be directed to magemberling "at" davidson.edu.