Tutorial Overview
This tutorial is designed to teach someone with limited computer programming experience how to utilize the power of local database blasting in their own programs. MacOSX is used in the tutorial, however, the use of standalone blast does not differ significantly across operating systems. In order to get the most out of the tutorial, it is best to have at least some knowledge of how to use command line applications, although this is not necessary to progress through the steps below. For more on using the command line, you can go through the Unix Tutorial for Beginners.Why is Standalone BLAST useful?
In the Genomics Lab class at Davidson College, I was given the task of creating a tool that assisted in our mission of investigating pathways in the Halorhabdus utahensis genome. Specifically, this tool was intended to help determine whether a given EC number existed in the genome but had not been annotated as such. To do this, I created a tool that, given an EC number, would find all known protein sequences with that EC number and blast them against the predicted proteins of our organism. If hits were found, this was suggestive of the given EC number existing in our genome. The tool I created can be viewed here and the perl program can be downloaded here.In order to make this tool, I needed to be able to blast known protein sequences against the Halorhabdus utahensis genome. The best way to do this was to run a local blast search on the genome. The tutorial below explains how this can be accomplished.
Downloading BLAST
To get the files you will need, go to the archives section of the BLAST database: http://www.ncbi.nlm.nih.gov/BLAST/download.shtmlChoose the appropriate blast download for your operating system. MacOSX users should choose the option highlighted in yellow.
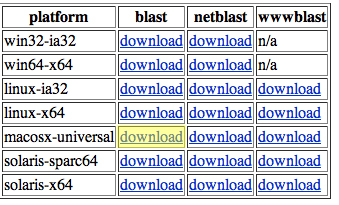
Download this file to your desktop and decompress it. You should have a folder entitled "blast-2.2.18" on your desktop. This folder contains all of the necessary executables (programs) in the "bin" subfolder. For more information on this download, including detailed descriptions of each executable, visit NCBI's documentation page.
Creating a new database
The first thing that we want to try to do is create a blastable database out of a preexisting file. For the purposes of this tutorial, let's imagine that you have a fasta formated file of amino acid sequences (sample.faa) and you want to be able to blast any amino acid sequence against the contents of this file. Move this file to the data folder within the blast-2.2.18 folder.You now need to create a blastable database using the "formatdb" executable in the bin folder. This can be done easily on the command line. Open Terminal (or another command line application) and navigate to the blast folder (Tutorial Note - command line entries will be highlighted in green):
cd Desktop/blast-2.2.18/
You will now need to run the formatdb executable. To list the command line options for this executable you can enter:
./bin/formatdb -
We will need to specify 3 arguments when running formatbd: -i, -p, and -o. -i specifies the location of the fasta formated sequence file, which in our case is "data/sample.faa." -p asks whether the sequence file contains protein sequences (versus amino acid sequences). Since we have a protein sequence file, this argument will be set to T (for true). Had it have contained nucleotide sequences, we would have set it to F. Finally, we have to set the parsing options, -o. For the purposes of this tutorial it is enough to simply set them to F (false). For more complex functionality within the database, you could set it to true. With these arguments in mind, enter the following command to generate a blastable database out of the sample.faa file:
./bin/formatdb -i data/sample.faa -p T -o F
If you look in the data subfolder of the blast-2.2.18 folder, you will see that 3 new files have been added: sample.faa.psq, sample.faa.pin, and sample.faa.phr. These are the database files that will be used when running a blast on the sample.faa sequences.
You can obtain additional fasta formatted files for individual organisms by searching GenBank for an organism's genome. Downlad the fasta formatted file by selecting the "file" option under the "send to" drop down.
If you want to perform blasts on the large databases used by NCBI's BLAST (such as the entire nucleotide collection, nr) you can download those databases locally by visiting NCBI's documentation page and clicking on the BLAST databases link at the bottom. Note that these files are often over 1 GB in size so the download may take a while.
Running a local blast search
Now that I have explained how to create a blastable database locally, I will discuss how to perform a blast of the sample.faa database. This can be done easily from the command line.First you must open Terminal again and navigate to the desktop directory:
cd Desktop/
You also need to create a fasta formattted file containing the sequence or sequences that you want to use as your query. For this tutorial, I will use the query file query.faa. Place this file on your desktop. If you list the files in your current directory using the li command, query.faa should appear.
The blast executable that we are going to run is called blastall. It is located in blast-2.2.18/bin. To see the arguments for this executable, enter:
./blast-2.2.18/bin/blastall -
We will need to include 5 arguments in our command to actually run the blast. -p sets the program to run. Since we are using protein sequences, we will run a blastp. -i gives the file name of the query sequence(s), query.faa. -d gives a path to the database file which in our case is located at blast-2.2.18/data/sample.faa. -e gives the e-value which we will set to .03. Finally -m determines the type of output. For full output, set this value to 0. For summarized output you can enter 9. Based on these arguments, enter the following command:
./blast-2.2.18/bin/blastall -p blastp -i query.faa -d blast-2.2.18/data/sample.faa -e .03 -m 9
This should give 5 separate blast hits from different sequences within the sample.faa file. Now run the file with -m set to 0 to see the full alignments:
./blast-2.2.18/bin/blastall -p blastp -i query.faa -d blast-2.2.18/data/sample.faa -e .03 -m 0
If you want to implement these commands in a perl program, you could do so by simply flanking it with single tick marks to indicate that it should be run from the command line. A piece of perl code would look something like this:
$output = `./blast-2.2.18/bin/blastall -p blastp -i query.faa -d blast-2.2.18/data/sample.faa -e .03 -m 0`;
In this case, $output would contain the output of the blast search.