
Read how scientists and journalists both portrayed this new genetic discovery in their respective articles.
This web page was produced as an assignment for an undergraduate course at Davidson College.
| Some Fact Concerning Alzheimer's | What the Scientists Discovered | How the Popular Press Conveyed It |
|---|
To understand Alzheimer's disease, scientists have been using genetics. Studies suggest that genes are the basis for more than 60% of Alzheimer’s susceptibility. They have found that mutations in three genes that explain the symptoms of early-onset Alzheimer’s, however these findings only help to explain about 1% of all Alzheimer’s. The APOE (apolipoprotein E) gene is often identified as a susceptibility locus for late-onset Alzheimer’s disease. Out of the previously stated 60%, 50% may be account for by APOE.
The researchers wanted to understand the random 10% and used genome-wide association studies (GWAS) to fulfill this knowledge gap.
They analyzed 537,029 SNPs in 2,032 French Alzheimer’s disease cases, diagnosed by neurologists, and 5,328 controls. All the samples were genotyped, for the first stage, with Illumina Human 610- Quad BeadChip. Using logistic regression, the resulting GWA data was further analyzed with the inclusion of sex, age at time of diagnosis, and possible population stratification adjustments.
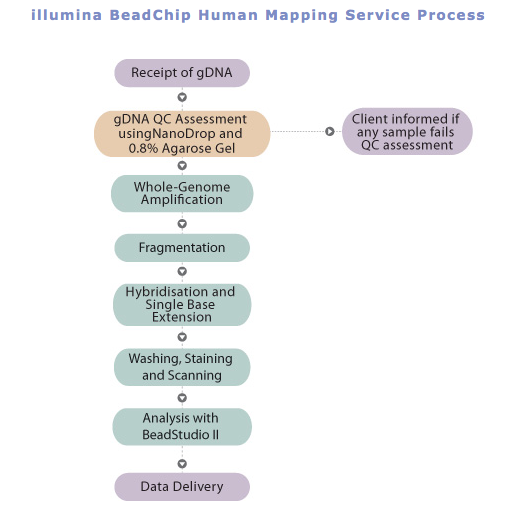
http://www.agrf.org.au/assets/images/Flow%20Charts/illumina-Whole-genome-SNP.jpg
Several APOE-linked (single nucleotide polymorphisms) SNPs strengthened this gene's association with the disease.
The second phase started off with a replication of association by genotyping selective loci as well as collecting more cases and controls from Belgium, Finland, Italy, and Spain. They found and genotyping them at the loci identified using Taqman (PCR) or Sequenom assays. Sex and age at diagnosis of whether they had dementia were factors in the data, which was analyzed using a logistic regression under an additive genetic model.
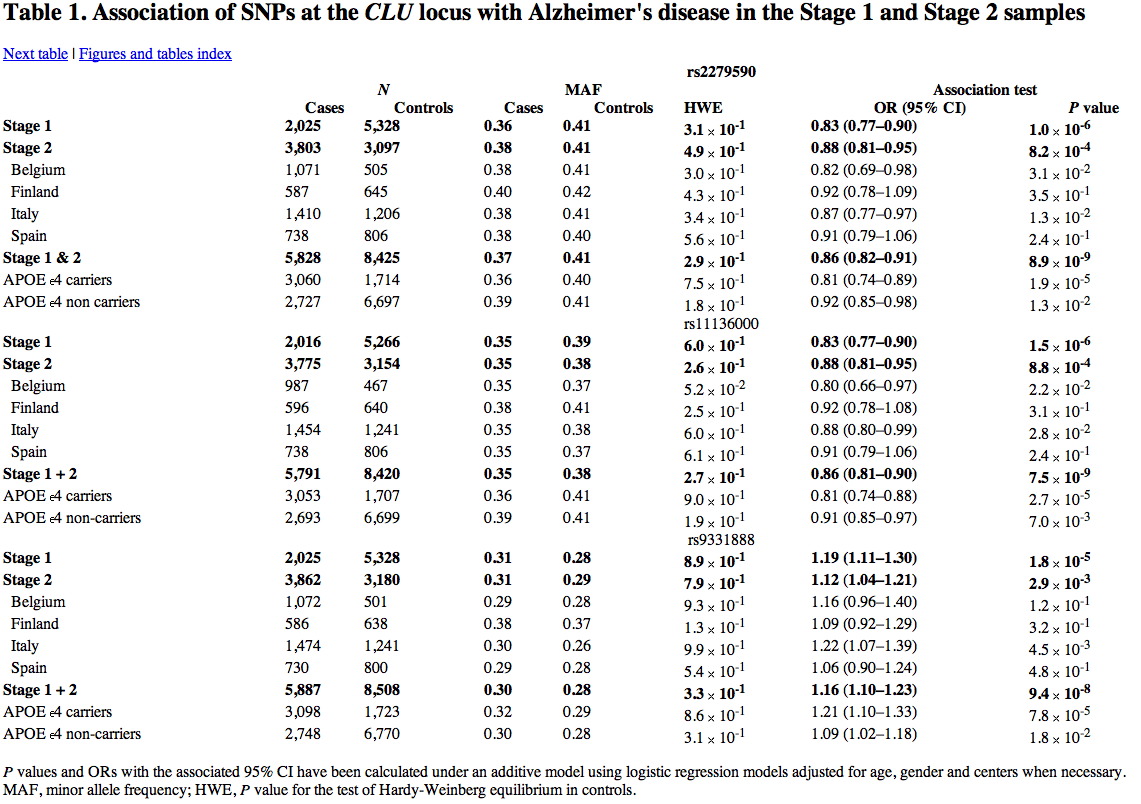
At the CLU locus, three markers (rs2279590, rs11136000, rs9331888) showed significance statistically, which emphasized CLU’s role in Alzheimer’s; also, a statistical interaction between APOE e4 status and the CLU SNPs. While all three markers show statistically significant association with Alzheimer's disease, one marker within CLU (rs11136000) on chromosome 8p21-p12 showed the most significant results in the association test with a P value of 7.5 x 10^-9. According to this article, the P values surpassed the criteria for genome-wide significance with the conserved Bonferroni correction (P< 4.5 x 10^-3), a statistical method that works to “correct” multiple comparisons when dependent and independent statiscal tests are being used concurrently. While the association was significant in both e4 carriers and non-carriers, it’s stronger with the carrier version of APOE.
They found that three CLU locus markers replicated in the second phase were within a linkage disequilibrium block, meaning that these markers were associated with each other non-randomly. The haplotypes that they define accounted for 98.2% of the observations in stage 1 controls. Their biological function as well as other markers from that region, is currently unknown.
Other markers were observed in many other chromosomes, one such marker was CR1.
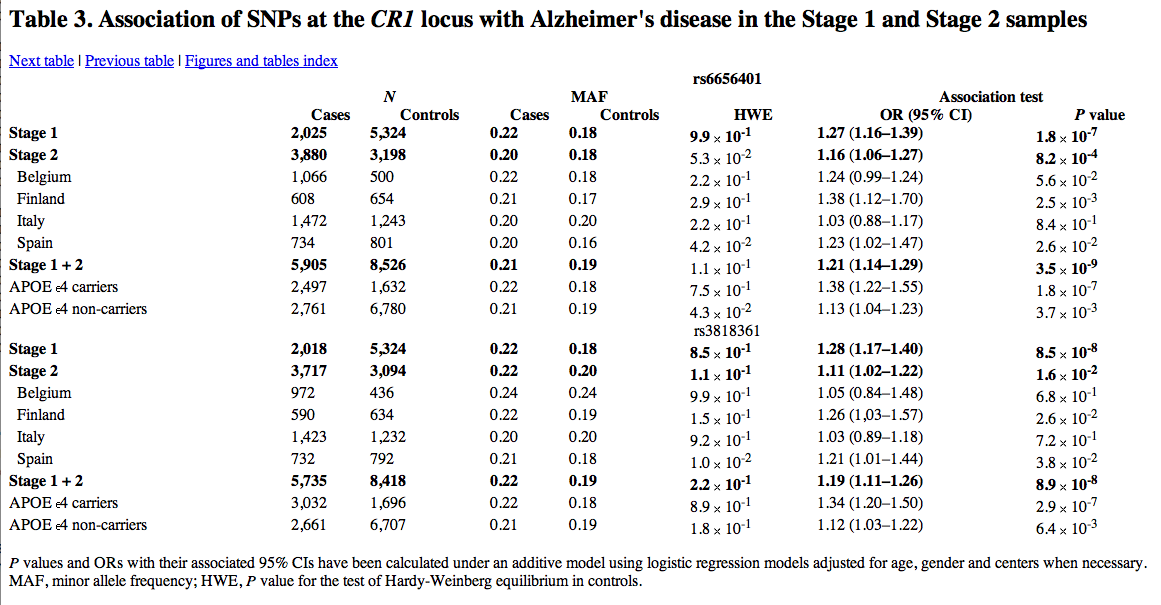
The linkage disassociation block that encompasses CR1 on 1q32 had one SNP (rs6656401) that had evidence of being associated with Alzheimer’s disease. At stage 2, the P value is 8.2 x 10^-4 however at Stage 1 the P value is 1.8 x 10^-7. Yet, there was no significant difference between their origins. At this locus, they found a statistical interaction with APOE e4 status and disease risk, P-value of 9.6 x 10^-3. Again, there was significant association with e4 carriers and non-carriers, with a stronger association with e4 carriers. Association to APOE e4 of the second marker at rs3818361 only showed significance in stage 2 data.
PILCAM, another gene, and its markers have also been found to be associated with late-onset Alzheimer's disease as found in the second study, although some association was found in the first study. However, the publication doesn't elaborate much more on this gene but results are online in GWAS results.
Conclusions regarding the biological roles of CLU and CR1
APOE and CLU are both apolipoproteins in the central nervous system that are the most expressed, and may have a huge impact on the physiopathological process of Alzheimer’s disease. CLU is present in amyloid plaques like APOE and can bind ABETA, the peptide that accumulates to form the plaque. Experiments with transgenic mice that were lacking CLU showed that this protein might be involved in vivo in ABeta fibrillogenesis and increasing ABeta neurotoxicity. Another proposal is that is may function like APOE in clearing ABeta from the brain across the blood-brain barrier.
CR1 is the gene that encodes the main receptor of the complement C3b protein. Through CR1, C3b mediates phagocyotosis after covalently binding to acceptor molecules. C3b is a fragment of C3 after it has been cleaved. The biological function of CR1’s markers is unknown but the linkage disassociation block only includes this gene, which proved to be statistically significantly associated with Alzheimer’s disease. Observations suggest that this gene-protein pair are involved in ABeta clearance. The reason for this, besides that this mechanism is associated with Alzheimer’s and with the expression of the gene/protein pair, are that ABeta accumulated can bind to C3b, an increase of C3 expression coincides with less ABeta neuropathological effects in a transgenic mouse while C3 convertase inhibitor results in more ABeta and neurodegeneration. In human red blood cells (the cells that expresses CR1 the most), the scientists saw that these cells were able to appropriate ABeta and favor its clearance when C3b bound to CR1.
All in all, these loci have been identified to be associated with ABeta clearance along with APOE. Early-onset is mainly associated with genes causing ABeta abundance, while the genetic variants at the CLU, CR1 and APOE loci most likely influence late-onset Alzheimer’s disease due to their roles in ABeta clearance.
The scientific article was definitely written for scientists with statistical background and was only somewhat comprehensive at the end. The infusion of statistical evidence into the text made the paper difficult to understand everything and absorb the information. Many of the techniques used were very advanced techniques that require lots of outside knowledge that even a biologist, non-genomist probably would not know about. The paper was very dense and only really showed insight at the end. While the evidence of whether these two genes associate with APOE is extremely significant. It could have been more concise and friendly towards non-genomists.
The popular press did a very poor job of explaining what this paper was trying to say so the general public had a very skewed vision of what the research represented.
Reference
Lambert, J. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease [Internet]. Nature Genetics. Published online: 6 Sept 2009 [cited: 14 Sept 2009] Available from: http://www.nature.com/ng/journal/vaop/ncurrent/abs/ng.439.html