AUTOMATED FLUORESCENT DYE-TERMINATOR CYCLE SEQUENCING
In 1980, Sanger devised the chain-termination dideoxynucleotide method for sequencing. The fluorescent dye-terminator cycle sequencing method is based on this concept, and corporations such as Perkin-Elmer, division Applied Biosystems Inc., have designed an automated method that combines the Polymerase Chain Reaction(PCR) with the actual sequencing in a sophisticated computer program(Fig1).

The PCR reaction includes the following ingrediants: DNA template, primer, Taq DNA polymerase, unlabeled dNTP's, fluorescently labeled ddNTP's, buffer, and distilled water. The fluorescently labeled ddNTP's are called dideoxynucleotides, named so because of their structure(Fig 2). Because they lack a 3' OH, nothing can be added to the chain once a ddNTP has been added; therefore, the sequence terminates in a fluorescently labeled ddNTP. Each base has a differently colored dye that can be recognized by a laser.
Fig. 2: top: deoxynucleotides used in normal PCR; bottom: dideoxynucleotides used for sequencing--labeled with a fluorescent dye. <http://www.plattsburgh.edu/acadvp/artsci/biology/bio401/DNASeq.html
During PCR, the temperature is increased to ~95 degrees Celcius to denature the double-stranded DNA. Then the temperature drops to ~50 degrees Celcius to allow the primers to anneal to the template. Finally, Taq polymerase adds nucleotides to the growing chain at ~60 degrees Celcius. This cycle repeats 25-30 times. During the last step, the Taq polymerase adds either the dNTP's or the ddNTP's to the chain randomly. Therefore, when PCR is complete, you are left with PCR fragments of all different lengths; for whenever a ddNTP was added, the elongation stopped.
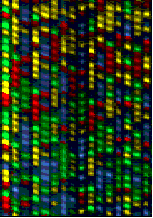
You can then load the entire PCR reaction into one lane of a polyacrylamine gel in a special laser sequencer(for example, the ABI 377). The sequencer can read the different dyes as they pass the laser at the bottom of the gel. Because the different fragments migrate toward the positively-charged pole at different rates, the shorter fragments migrate toward the bottom of the gel faster, so you read it bottom->top(5'->3'). The computer then compiles these data into the image of a gel(Fig. 3). The colors are then read as bases to produce a chromatogram of the piece of DNA sequenced(Fig. 4). The sequence can be taken right from the chromatogram with an extremely high level of accuracy, and the task is complete!
 5'
5'
Fig. 3: An actual polyacrylamide sequencing gel with 16 lanes, each loaded with a different sequence sample. Each one of the four colors represents a different base. By translating the colors to bases, you can determine the sequence of the DNA that PCR amplified. <http://www.sci.sdsv.edu/dnacore/overview.html>
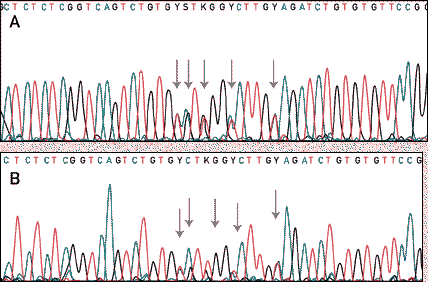
Fig. 4: A chromatogram of the DNA sequence. The computer reads the colored peaks as one of four bases, translating only at strong peaks. 'Y' represents a base that was ambiguous--either there were two conflicting signals or there was no signal when the computer was expecting one. The gray arrows show where the computer would expect to read a base. <http://www2.perkin-elmer.com:80/ga/uu0901/770901.html>
To return to my homepage, click here.