*This website was produced as an assignment for an undergraduate course at Davidson College.*
ATP Synthase:
A vital protein used in oxidative phosphorylation

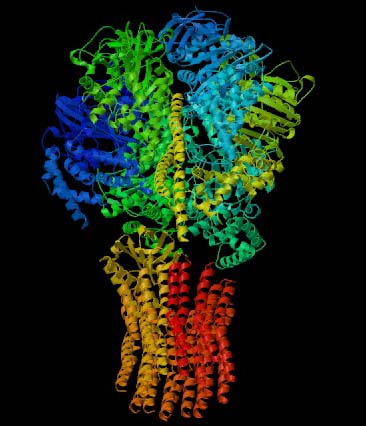
Figure 1. This 3-D image of ATP Synthase illustrates the protein's complex tertitary structure. Subunits are designated by different colors. Permission for use of this image is presently being sought from the site <http://www.blc.arizona.edu/courses/181gh/rick/photosynthesis/F1-ATP_synthase.html>, where the image can be found in its original context.
This webpage explores the structure and function of this interesting protein, as well as the medical consequences of malfunctioning ATP Synthase. I have also included links to related sites.
What is ATP Synthase?
ATP Synthase, also known as Complex V, is a large membrane protein that plays a key role in the energy metabolism of all known organisms (Nowroth, 2003). Located in the inner membrane of the mitochondria, ATP Synthase is involved with the electron transport chain within the same membrane. The 4 complexes of the electron transport chain create a concentration gradient of protons across the membrane; ATP Synthase is then able to utilize this concentration gradient to produce ATP. Figure 2 below gives a good overview of the flow of electrons through the respiratory chain (Complexes I-IV) and then ATP Synthase's ability to produce energy for the cell in the form of ATP.
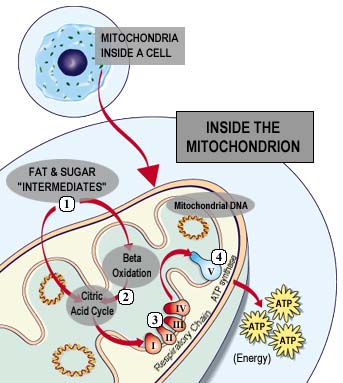
Figure 2. This image illustrates the location of oxidative phosphorylation and ATP Synthase in the cell and also provides a general idea of how the enzyme is able to produce ATP for the cell. Permission for use of this image is being sought at <www.mitoresearch.org/Quest_6_4.htm>, where it can be found in its original context.
According to the chemiosmotic model proposed by Peter Mitchell, a proton-motive force is created across the inner membrane, derived from the electrochemical energy inherent in the difference in proton concentration and separation of charge across the membrane. This proton-motive force drives the synthesis of ATP as protons flow passively back into the matrix through ATP Synthase (Nelson et al., 2000). Since the inner mitochondrial membrane is impermeable to protons, the proton-specific channel of ATP Synthase (Fo) is the only route for protons to reenter the matrix. To understand this process in detail, we must analyze the protein's structure.
Let's take a closer look at the structure of ATP Synthase...
ATP Synthase is an F-Type ATPase, one of the four categories of ATPases; all ATPases in this category share the central role of the catalytic formation of ATP from ADP and Pi . All F-Type ATPases also have a peripheral domain (F1) and an integral domain (Fo) and pump protons only in one direction (Nelson et al., 2000).
Figure 3 clearly highlights Fo and F1 and illustrates the many different subunits of this complex--ATP Synthase has a total of > 20 protein subunits in most organisms! John E. Walker and colleagues determined many structural details of the enzyme using crystallographic and biochemical studies. According to this model, F1 has nine subunits of five different types; three beta subunits and three alpha subunits are arranged like the segments of an orange, with alternating alpha and beta for a total of six subunits. The knoblike portion of F1 is a flattened sphere 8 nm high and 10 nm across (Nelson et al., 2000). A central shaft (gamma) runs through the center of these six subunits, as an alpha-helical coil. The delta and epsilon subunits are attached to the gamma subunit and help link the F1 domain to the Fo domain. This central stalk rotates at about 50-100 times per second (Walker, 2002). Figure 4 shows a structural view of F1 from below.
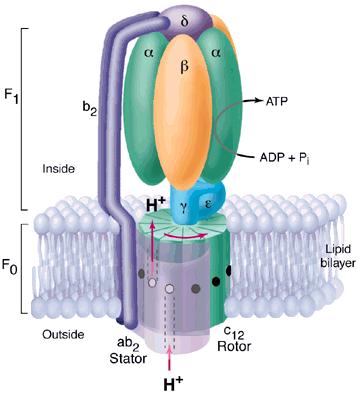
Figure 3. This diagram of ATP Synthase within the mitochondrial inner membrane shows two major strutural components Fo and F1 and the subunits within each. Permission for use of this image is being sought at <http://pathology.mc.duke.edu/research/postpizzo.html>, where it can be found in its original context.
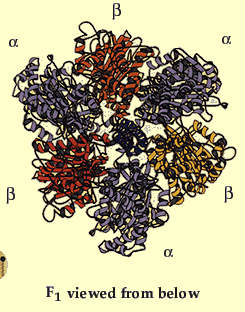
Figure 4. This image gives a better view of the arrangement of the F1 complex and the arrangement of the alpha and beta subunits around the central stalk. Permission for use of this image is being sought at <http://www.nobel.se/chemistry/educational/poster/1997/boyer-walker.html>, where it can be found in context.
As shown in Figure 3, the Fo portion consists of three subunits: a, b, and c. The two b subunits associate firmly with the alpha and beta subunits of F1, holing them fixed to the membrane. The membrane-embedded cylinder of c subunits is attached to the shaft of F1. As protons move through the membrane via Fo, the cylinder and shaft rotate, and the beta subunits of F1 change conformation as the gamma subunit associates with each in turn (Nelson et al., 2000).
How do the conformations of the Beta subunits of F1 change?
The amino acid sequences of all three beta subunits are identical, but their conformations differ, mainly because of the association of the gamma subunit with just one of the three. The ATP-ADP binding sites of the three beta subunits differ and are labelled beta-ATP, beta-ADP, and beta-empty. This difference in binding is critical to the protein's mechanism. For every three protons that flow through ATP Synthase, the complex rotates one position (due to the rotation of the gamma subunit) and one ATP molecule is formed. The three beta subunits interact in such a way that when one subunit releases ATP and become beta-empty, its neighbor to one side must assume the beta-ADP conformation and the neighbor one the other side must assume the beta-ATP conformation. Thus, for each complete rotation, three ATP are synthesized and released (Nelson et al., 2000). The image below shows this rotaion.
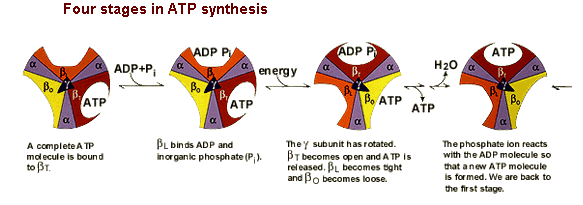
Figure 5. This image shows the conformations of the beta subunits as the complex rotates. For every 3 protons that go through the enzyme, one ATP is produced and released. This image was found at the same site as Figure 4 and permission is currently being sought.
Orthologs of ATP Synthase
As mentioned before, there are four classes of ATPases and ATP Sythase is an F-Type ATPase. The structures and functions of F-Type ATPases and all four types of ATPases differ slightly among species. For example, ATP Synthase is found in the thylakoid membrane of higher plants and in the plasma membrane of prokaryotes (compared to the mitochondrial membrane of eukaryotes). In each of these cases, however, ATP Synthase functions to catalyze the formation of ATP from ADP and Pi (Nelson et al., 2000).
Furthermore, V-Type and F-Type ATPases in particular share common structural components. Like ATP Synthase, V-type ATPases have a peripheral domain (V1) and an integral membrane component (Vo). V1 has seven different subunits, with three A and three B around a central stalk D. Vo also has three types of subunits with muliple copies of subunit c. Obvioulsy this structure is very similar to that of F-type ATPases, but V-type ATPases are located in vacular membranes of higher plants and funghi and in lysosomal, endosomal, and secretory vesicle membranes of animals rather than the mitochondrial inner membrane (Nelson et al, 2000).
ATP Synthase Mutations
There is evidence that NARP syndrome is caused by a point mutation at nucleotide 8993 on the gene encoding subunit 6 of mitochondrial ATP Synthase. In a 1990 study, there was correlation between clinical severity and the amount of mutant mitochondrial DNA in patients. The symptoms included developmental delay, retinitis pigmentosa, dementia, seizures, ataxia, proximal neurogenic muscle weakness, and senory neuropathy, in a petigree consistent with maternal transmission (Kelly, 2000).
Prior to a 1999 report by Houstek et al., all known ATPase defects were caused by maternally inherited mtDNA mutations in subunit 6 of the Fo complex (one of the two Fo subunits that are products of mitochondrial genes). But Houstek et al. described a new selective defect of mitochondrial ATP Synthase caused by altered biosynthesis of the enzyme. This defect manifests itself as fatal neonatal lacticacidemia with cardiomegaly and hepatomegaly (McKusick, 1999).
In general, any mutation or poison that causes loss of function of ATP Synthase proves fatal because gylcolysis alone does not produce enough ATP to sustain life. Oligomycin and venturicidin are both examples of toxic antibiotics known to bind ATP Synthase and consequently inhibit ATP synthesis (Nelson et al., 2000).
Links to Related Sites
Here is a link to a lecture with many great images--definitely check it out!
<www.life.uiuc.edu/crofts/bioph354/lect10.html>
This site gives a good overview of oxidative phosphorylation and ATP Synthase's role:
<www.gwu.edu/~mpb/oxidativephos.htm>
This link is an excellent reference for the general characteristics and classifications of human ATP Synthase, including its amino acid sequence, molecular weight, and database cross-references:
http://srs.sanger.ac.uk/srsbin/cgi-bin/wgetz?-e+%5bSWISSPROT-id:ATPO_HUMAN%5d#Links
References
Heller HC, Orians GH, Purves WK, Sadava D. Life: the Science of Biology, sixth edition. Sunderland, Massachusetts: Sinauer Associates, Inc., 2001. 319.
Kelly, J. Narp Syndrome (#551500). Accessed on OMIM database, 2003 March 10.
McKusick, VA. ATP Synthase Deficiency (#604273). Accessed on OMIM database, 2003 March 10.
Nelson, DL, Cox MM. Lehninger Principles of Biochemistry, third edition. New York: Worth Publishers, 2000. 675.
Nowroth, Dr. Thomas. Membrane protein structure dynamics: ATP Synthase. <http://mpsd.de/ATP-Synthase.html> Accessed 2003 March 10.
Walker, JE. ATP Synthase. <http://www.mrc-dunn.cam.ac.uk/research/atpase.html> Accessed 2003 March 10.
Please direct questions or comments to Caroline Bennett at cabennett@davidson.edu. Thank you for visiting my site!