This web page was produced as an assignment for an undergraduate course at Davidson College
Phage Display
Purpose
The main purpose of phage display is to determine proteins that will interact with disease causing molecules with the intent of disarming these molecules and curing or weakening the disease.
What is a phage?
A bacteriophage (phage) is a virus that infects only bacteria. The two major components of a phage are the genetic material and the protein coat. When the phage enters a cell its DNA is expressed causing more phages to be made. The purpose of the protein coat is to protect the genetic material from being damaged as the phages reproduce and go from cell to cell.
Phages used in phage displays are normal phages that have been genetically modified to express one, and only one, extra protein that will be on the exterior of the phage (see Figure 1).
Figure 1: This is a cartoon of a phage used in phage displays. The blue portion on the left side of the phage represents the extra protein that is expressed by this phage. (click on image for source).
Phage Displays: The Process
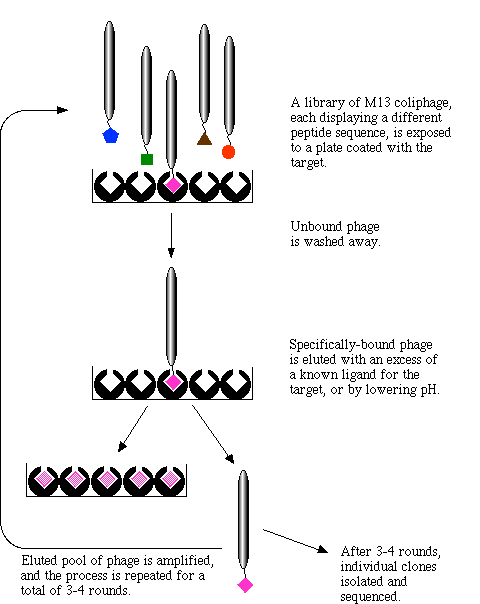
1. Insert a diverse group of genes into the phage genome. Each phage recieves a different gene. Generally, the gene is inserted into the gene 3 position so that the protein expressed will be located on the surface of the phage. Since phages are so small, as many as a billion proteins can be inserted, each with its own phage.
2. Create a library of genetically modified phages which are all related but each of which have a different gene from step 1.
3. Isolate a disease causing molecule, such as a receptor or an enzyme, and expose the phage library to the molecule.
4. Some of the phages will bind to the disease causing molecule because there is an interaction between it and the protein inserted earlier. Wash the phage library. All of the phages with proteins that didn't bind to the disease causeing molecule will be removed.
5. Replicate the phages that remain so that they can be sequenced.
6. Since we know where the new gene was inserted we can determine the amino acid sequence of the protein that bound to the disease causeing molecule (Dyax, 2003, <http://www.dyax.com/phage/phage.asp>).
This cartoon (Figure 2) describes the process from step 2 through step 6:
Figure 2: This cartoon describes the process of phage display. The depiction begins with a phage library where each phage is expressing a different protein that has the potential to interact with a disease causing molecule. Click on image for source.
Applications
The main practical application of phage displays is to use the drugs that have been isolated by this simple process to design drugs that either cure or curtail the effects of specific diseases. Further investigation needs to be done once the phage display process is complete to determine the specificity with which the protein binds as well as other qualities of the protein that might make it a good drug candidate. However, this process allows researchers to zoom in on the few proteins that actually interact with disease causing molecules instead of searching for any protein that might function well as a drug. In effect, phage display helps researchers to engineer human antibodies. Other applications include arrays and separations (Phage Display Technologies, 2002, <http://www.healthtech.com/2001/pgd/>).
Phage libraries can be constructed in weeks alone and screening takes only a few extra days. This is much more time efficient than older methods that attempted to accomplish the same goals(Dyax, 2003, <http://www.dyax.com/phage/phage.asp>).
Key Elements
Phage displays utilize serveral key biological processes. The first, is the idea that when two proteins bind together, there is a change in the configuration of the proteins often causing a protein to change its function or not function at all. This method identifies proteins that bind to disease causing molecules but without this proven biological principle that protein binding changes protein shape, the method would be ineffective (Phage Display Technologies, 2002. <http://www.healthtech.com/2001/pgd/>).
The effectiveness of phage displays depends heavily on the comprehensiveness of the phage library that is used. Naturally, if only a few proteins are included in the library, the phage display will yeild limited results. Although there are only 20 amino acids, there are a multitude of proteins out there, so investigators should work to create more and more complete phage libraries so that this pocedure can be utilized to the fullest (Dyax, 2003, <http://www.dyax.com/phage/phage.asp>).
Links to Related Sites
To learn more about phage displays and to see some practical applications of phage displays in science, check out these links:
http://www.dyax.com/phage/phage.asp
http://www.immun.lth.se/TEXTER/MO/phage_display.html
http://baserv.uci.kun.nl/~jraats/links1.html
References
Collins, J., Rottgen, P., Evolutives Phage-Display, eine Methode als Grundlage eines neuen Unternehmens, 2002, <http://bib.gbf.de/ergebnisbericht/1997/deutsch/sektion_b/b-collins.html>
Combinatorial Development Of Peptide Tags For Linking Proteins To Synthetic Materials, 2002, <http://www.chem.tue.nl/smo/MBE/PhageDisplay.htm>
Dyax, 2003, <http://www.dyax.com/phage/phage.asp>
Ohlin, M., Antibody Phage Display, 2003, <http://www.immun.lth.se/TEXTER/MO/phage_display.html>
Phage Display and Antibody Engineering Techniques, Department of Biochemistry, University of Nijmegen, 2003, <http://baserv.uci.kun.nl/~jraats/links1.html>
Phage Display Technologies: Directed Protein Evolution, 2002, <http://www.healthtech.com/2001/pgd/>
This page was designed by Graham Watson, a senior Biology major at Davidson College.
Click here to go back to my Home Page
© Copyright 2003 Department of Biology, Davidson College, Davidson, NC 28035
Send question to grwatson@davidson.edu