GCAT Best Practices Workshop
August 13 - 15, 2003
Institute of Systems Biology in Seattle

Raw Data From Workshop - Each Tiff File is 22.5 MB
Slide Number |
Labeling Method |
RNA Samples |
Cy5 (red) Low Signal |
Cy3 (green) Low Signal |
Cy5 (red) High Signal |
Cy3 (green) High Signal |
| 4152 | Indirect Dye | Cy3 0.6: Cy5 4.0 | 4152_Cy5L.tif | 4152_Cy3L.tif | 4152_Cy5H.tif | 4152_Cy3H.tif |
| 4153 | Direct Dye | Cy3 0.6: Cy5 4.0 | 4153_Cy5L.tif | 4153_Cy3L.tif | 4153_Cy5H.tif | 4153_Cy3H.tif |
| 4154 | Direct Dye | Cy3 4.0: Cy5 0.6 | 4154_Cy5L.tif | 4154_Cy3L.tif | 4154_Cy5H.tif | 4154_Cy3H.tif |
| 4155 | Direct Dye | Cy3 0.6: Cy5 0.6 | 4155_Cy5L.tif | 4155_Cy3L.tif | 4155 Cy5H.tif | 4155_Cy3H.tif |
| 4156 | Direct Dye | Cy3 4.0: Cy5 4.0 | 4156_Cy5L.tif | 4156_Cy3L.tif | 4156_Cy5H.tif | 4156_Cy3H.tif |
| 4157 | 3DNA | Cy3 4.0: Cy5 0.6 | 4157_Cy5L.tif | 4157_Cy3L.tif | 4157_Cy5H.tif | 4157_Cy3H.tif |
Genelist for these data (.txt file)
Excel Version with column headings
Addressing these data
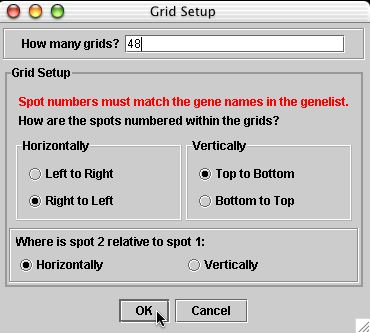
There are 24 columns and 23 rows in each grid.
There are 4 metacolumns and 6 metarows in the top half and this is duplicated again in the bottom half. Therefore, if you wanted to grid every spot no this microarray, there would be a total of 48 grids each composed of 24 x 23 arrays of spots.
Click to see a larger version of this image.
The experiment was designed to duplicate the DeRisi diauxic shift paper (1997). All of the RNA was prepared by Dr. Laura Hoopes' lab from two populations of cells. One group of cells was harvested at an optical density of 0.6 and the other was harvested at an optical density of 4.0. RNA was isolated and cDNA generated using one of three methods (indirect, direct, or 3DNA). Our goal was not to rate the three methods, but to determine if they worked and what pros/cons each method has. All methods did produce strong signals, so we are comfortable with GCAT members using the one they like best. It may be equipment, number of steps, time constraints, costs, etc. But GCAT does not want to tell people which method to do. We just want to make each method possible and to maximize your success.
The "Indirect Dye"method uses amino allyl dUTP to label the cDNA. This method prevents the dyes from creating a bias in the cDNA synthesis. The "Direct Dye" method uses Cy3/Cy5-coupled dUTP for cDNA synthesis. This reduces the number of steps to make the labeled cDNA. The 3DNA method uses Genisphere's dendrimer with about 200 dyes per cDNA so the signal is very strong. You can download the methods we used, and personal comments from the table below:
Tips we learned that help for all three microarray methods
Frequently Asked Questions About Microarry Results (work in progress)
Analyze the Data with MAGIC Tool
The Workshop
We heard talks by Lee Hood, John Aitchison, and Krassen Dimitrov from ISB. Then we made our plans and discussed curriculum during incubations.




We broken into three teams: wet lab (left), data analysis (center) and assessment
(right).



We had marvelous help from Steve who taught us a lot of methods, subtle tips, and a rich vocabulary.


Of course, the reason we were there....

Good data and good times.

