This web page was produced as an assignment for an undergraduate course at Davidson College.
Preface: This web page looks at gene expression patterns for KGD1 and YIL127C in yeast. The primary goal of this web page is to illustrate how DNA microarrays correlate gene expression patterns to function by the guilt by association method (Campbell et al., 2002). If a non-annotated gene's are expression pattern is similar to an annotated gene, then may share a common function, promoter or transcription factor.
From my second web project, I know the following information
about KGD1:
Biological Process:
The KGD1 gene encodes for 2-oxoglutarate dehydrogenase, a subunit in the alpha-ketogluterate
dehydrogenase complex, which is responsible for the decarboxylation of alpha-ketogluterate
in the TCA cycle. This enzyme plays an integral role in cellular respiration
and the production of ATP since its enzymatic reaction is coupled to the reduction
of NAD+. The enzyme also plays an integral role in amino acid metabolism.
Molecular Function:
The alpha-ketogluterate dehydrogenase complex is responsible for the catabolic
metabolism of alpha-ketogluterate to succinyl-CoA and CO2. The reaction requires
the cofactor thiamine pyrophosphate and the dehydrogenase complex consists
of three different subunits, 2-oxogluterate dehydrogenase (E1), dihydrolipoamide
succinyltransferase (E2) and lipoamide dehydrogenase (E3) encoded by KGD1,
KGD2 and LPD1 respectfully. The reaction also requires the cofactor thiamine
pyrophosphate.
Cellular Component:
2-oxoglutarate dehydrogenase and the alpha-ketogluterate dehydrogenase complex
localizes in the mitochondrial matrix. (SGD,
Swiss-Prot, (Mockovciakova, 1993))
Predictions: Before we analyze DNA microarray data, lets look at the metabolic pathways associated with KGD1 (see figure 1,2,3) These diagrams illustrate the enzyme interactions for yeast metabolism. If one component in a particular pathway is stimulated and expression is altered, then enzymes it interacts with should have similar expression patterns.
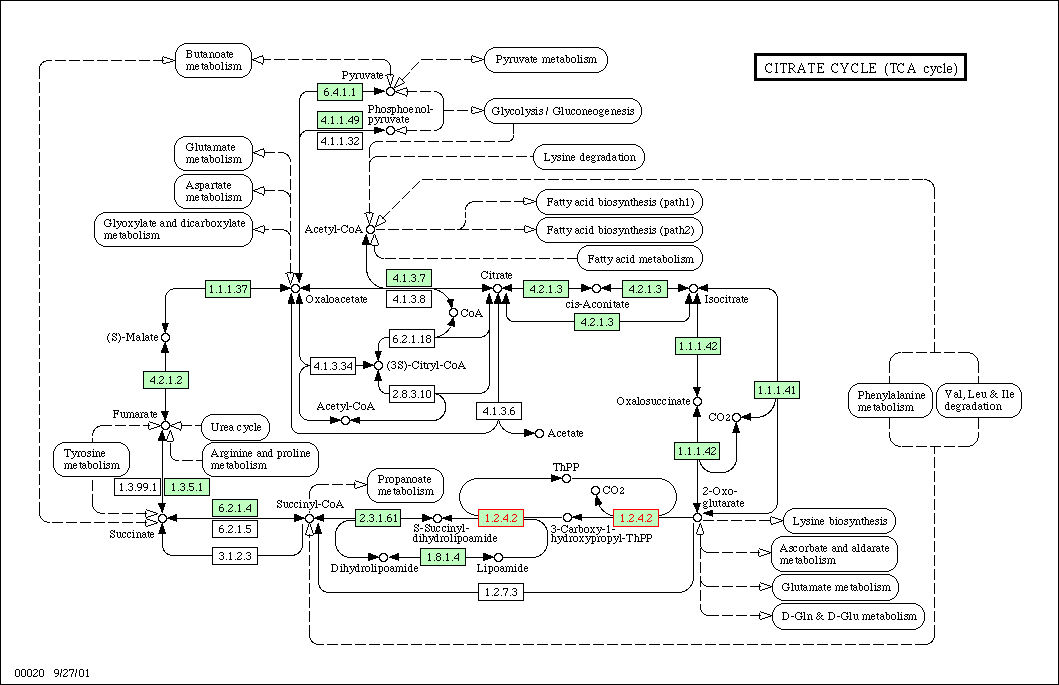
Figure 1: TCA cycle for yeast. KGD1 is enzyme 1.2.4.2. This enzyme works in conjunction with many different enzymes. One prediction is enzymes in this metabolic pathway would have similar expression patterns in different growth media.
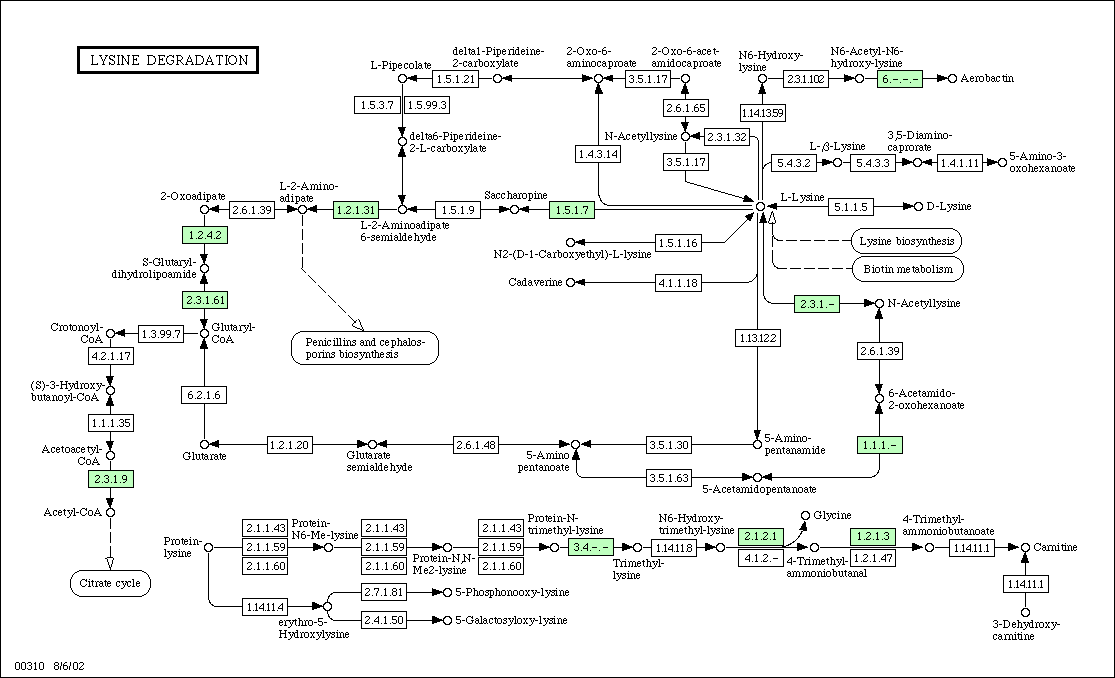 Figure 2:
Metabolic Pathway for Lysine
degradation in yeast. KGD1 is labeled as protein 1.2.4.2 and is located
in the upper left hand corner. For enzymes that regulate metabolism in association
with KGD1, one would predict similar expression patterns for DNA microarray
experiments.
Figure 2:
Metabolic Pathway for Lysine
degradation in yeast. KGD1 is labeled as protein 1.2.4.2 and is located
in the upper left hand corner. For enzymes that regulate metabolism in association
with KGD1, one would predict similar expression patterns for DNA microarray
experiments.
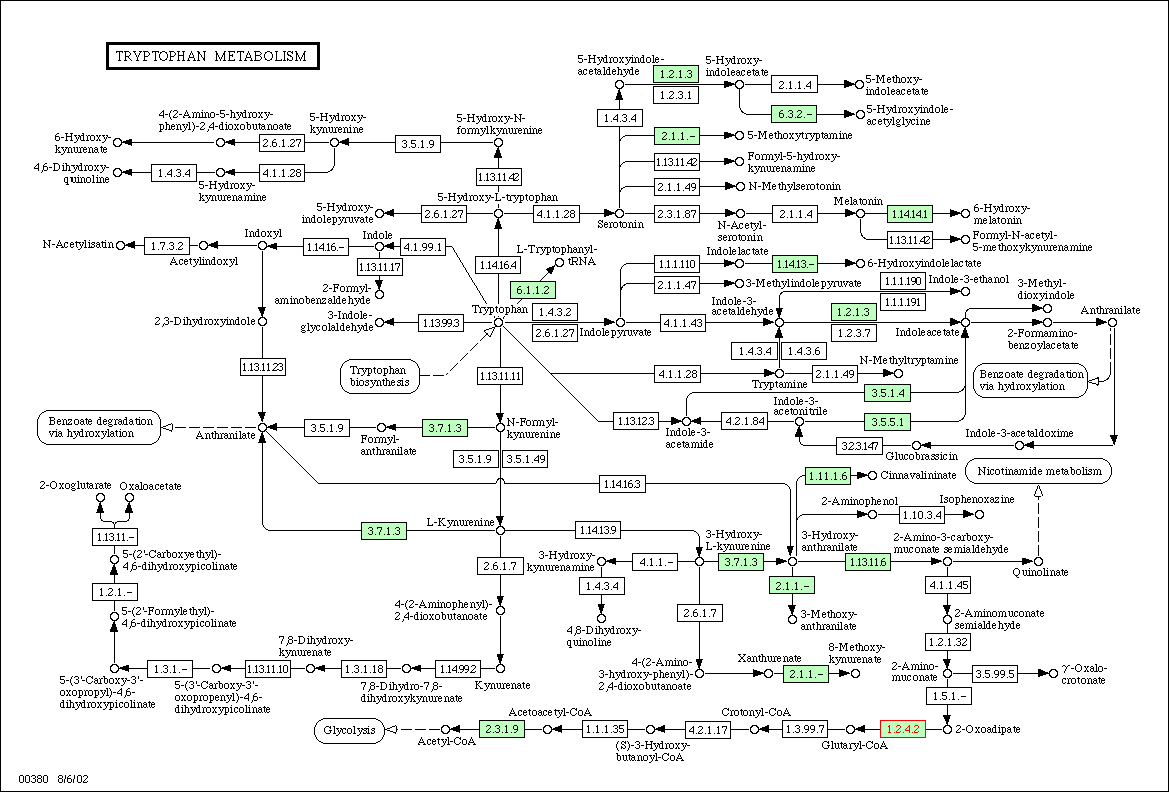 Figure
3: Metabolic diagram for tyrptophan
metabolism in yeast. The E1 subunit of the 2-oxoglutarate dehydrogenase
complex is located in the lower right hand corner and is labeled with red
text. Enzymes associated with 1.2.4.2 in tyrptophan metabolism should have
similar expression patterns.
Figure
3: Metabolic diagram for tyrptophan
metabolism in yeast. The E1 subunit of the 2-oxoglutarate dehydrogenase
complex is located in the lower right hand corner and is labeled with red
text. Enzymes associated with 1.2.4.2 in tyrptophan metabolism should have
similar expression patterns.
For YIL127C, database searches were inconclusive. The guilt by association method indirectly implies that genes localized in a chromosomal region have similar or interrelated functions because they are regulated by similar a transcription mechanism, i.e. they share a common promoter or enhancer. A database search at Swiss-prot revealed that genes surrounding KGD1 and YIL127C (see figure 4) the have the following functions:
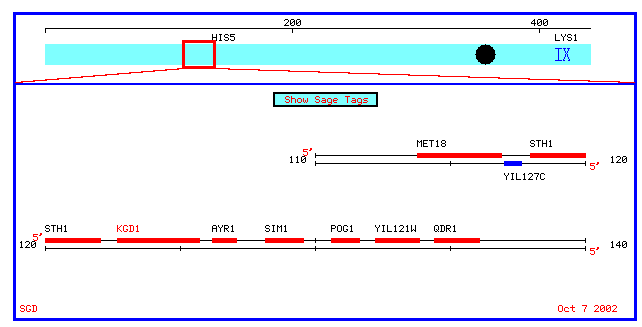
Figure 4: Physical map which spans 10 kb upstream and downstream of KGD1 on chromosome 9 (coordinates 112689 to 135733).
MET18:
Swissprot lists the function of Met18 as "INVOLVED IN
NUCLEOTIDE EXCISION REPAIR (NER) AND RNA POLYMERASE II (POL II) TRANSCRIPTION.
IT PROBABLY DOES NOT PARTICIPATE DIRECTLY IN NER AND POL II TRANSCRIPTION
BUT EXERTS ITS BIOLOGICAL EFFECTS BY INFLUENCING THE ACTIVITY OF TFIIH AND
POSSIBLY OTHER DNA REPAIR AND TRANSCRIPTION FACTORS AS AN UPSTREAM REGULATORY
ELEMENT. INVOLVED IN SPORULATION AND POSSIBLY IN THE RAD52 RECOMBINATIONAL
REPAIR PATHWAY." The subcellular location is nuclear.
STH1: This
is a nuclear protein "essential for mitotic growth and involved in the
G2 phase. The subcellular location is nuclear.
AYR1: Currently
there is no listed function at Swiss-Prot.
SIM1: Currently
there is no listed function at Swiss-Prot.
Notice that the upstream genes MET18 and STH1 have a nuclear subcellular location. These genes also do not play a role in metabolism. The relation between STH1 and MET18 may be that both genes encode for proteins that are needed during some part of the cell cycle.
My hypothesis is that if these genes are regulated by a common promoter, then I would expect to find some DNA microarrays that show that these genes have similar expression profiles or are repressed and induced simultaneously. Most of the aforementioned proteins are associated with mitotic growth, and cell cycle regulation. Therefore, I would suspect that YIL127C has a function associated with the cell cycle.
Why is the KGD1 located near genes involved in mitosis and the cell cycle? Imagine, that when the cell begins to grow or mitoticly divide it needs extra enzymes for energy production. Therefore, genes responsible for metabolism may be expressed in conjunction with genes needed during the cell cycle. Keep in mind that a negative result does not mean these genes do not share a common promoter. It simply means that an experiment has not been preformed that proves this theory. For all I known, every gene in this region shares a common promoter and each gene shares a common transcription factor. However, if each gene required a specific transcription factor, then expression patterns for the genes in this chromosomal region would not be similar. Note: I have assumed that changes in gene expression coincide with similar changes in protein translation. So if a gene is induced 4X, then protein translation should be induced 4X.
MICROARRAY DATA FOR KGD1
Eleven different experiments are listed in the Expression Connection database:
Expression response to different alpha factor concentrations
(1)
Expression response to alpha factor (2)
Expression response to DNA damaging agents (3)
Expression response to diauxic shift (4)
Expression response to glucose limitation (5)
Expression regulated by PHO pathway (6)
Expression response to environmental changes (7)
Expression response during the cell cycle (8)
Expression response to histone depletion (9)
Expression response to sporulation (10)
Expression response to varying zinc levels (11)
Figure 5: Scale for gene induction and repression for all DNA microarray experiments.
Here are the results from my search. Each figure represents
a different DNA microarray experiment. 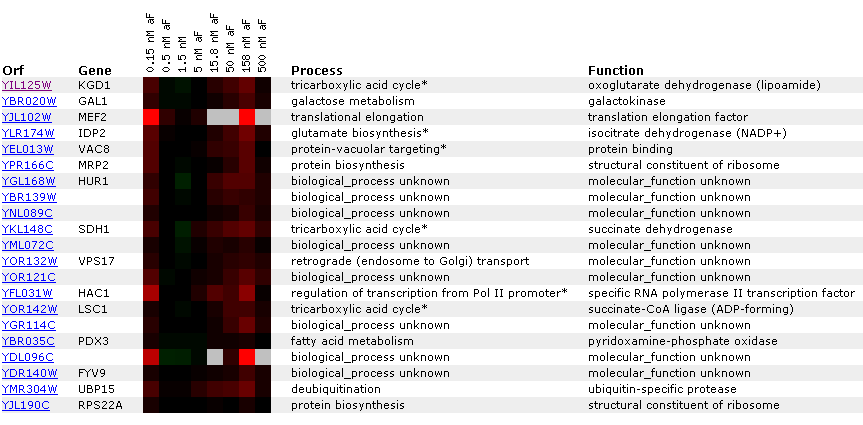 Figure
6: Gene
expression for KGD1 at various alpha factor concentrations. Genes listed
in the diagram have a Pearson correlation coefficient greater than 0.8. Notice
that KGD1 gene expression is slightly induced at 158 nM. For most alpha factor
concentrations the gene is barely induced. Notice that LSC1 and IDP2, proteins
associated with the tricarboxylic acid cycle have a similar expression pattern.
This suggests that enzymes in a metabolic pathway have similar expression
profiles.
Figure
6: Gene
expression for KGD1 at various alpha factor concentrations. Genes listed
in the diagram have a Pearson correlation coefficient greater than 0.8. Notice
that KGD1 gene expression is slightly induced at 158 nM. For most alpha factor
concentrations the gene is barely induced. Notice that LSC1 and IDP2, proteins
associated with the tricarboxylic acid cycle have a similar expression pattern.
This suggests that enzymes in a metabolic pathway have similar expression
profiles.
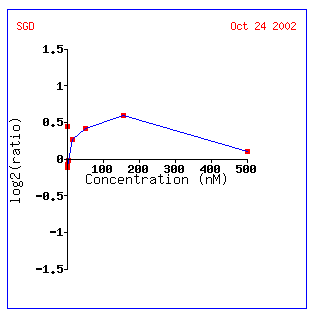
Figure 7: KGD1 expression profile for various alpha factor concentrations. KGD1 is slightly induced up to a maximum concentration of approximately 180 nM. This result may be indicative of a yeast's increased energy when it is stimulated by a pheromone.
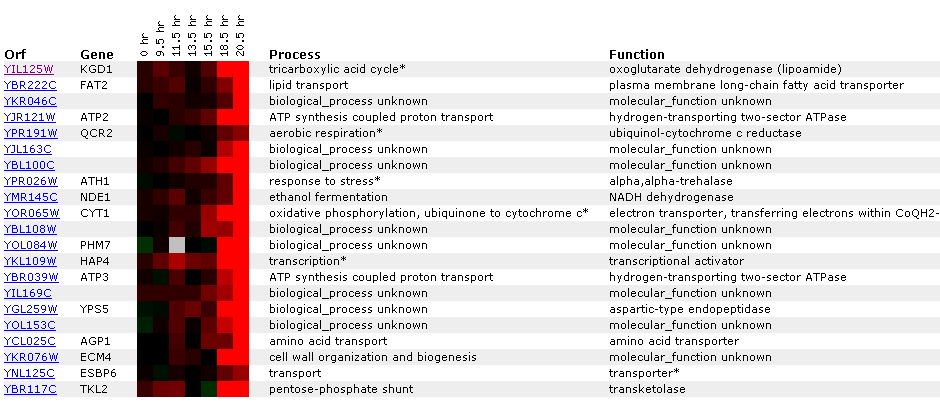
Figure 8: KGD1 expression during a diauxic shift. Notice that KGD1 is the only TCA cycle enzyme to be induced. KGD1 is slightly induced at 9-15 hours and induced greater than 2.8 fold after 18.5 hours. Gene's CYT1, ATP3, ATP2 and NDE1 are induced as well. These genes are responsible for oxidative phosphorylation, ATP synthesis, ethanol fermentation, respectfully. The microarray data indicates that the cell is producing more enzymes for aerobic metabolism. These results are expected since a diauxic shift results in switching from anaerobic metabolism to aerobic metabolism. ATH1 induction suggests metabolic pathway alteration requires large changes in the genome and that the transition from an anaerobic to aerobic environment is stressful to yeast. CYT1 has a similar expression profile to SDH1 (succinate dehydrogenase) and CIT1 (citrate synthase), genes associated with the TCA cycle(Data not shown). This implies, that TCA cycle genes may have similar expression profiles, but a correlation coefficient that is less than .8. Both SHD1 and CIT1 genes were induced at an earlier time point and maintained high induction levels for the remainder of the experiment.
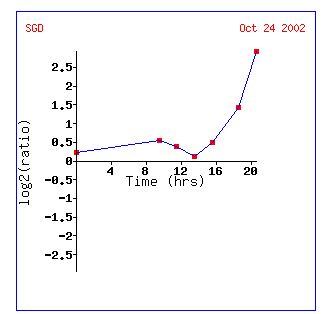
Figure 9: Gene expression profile for KGD1 for diauxic shift experiment. Expression increases significantly after 13 hours. This graph can be obtained by clicking on the link for diauxic shift in figure 8.

Figure 10: Comparison of transcriptional changes from parent strain to evolved strain with glucose limitation for 200 generations. KGD1 expression remains unchanged when comparing parent strain to evolved stain. KGD1 expression correlates to genes related to the cytoskeleton, and the cell cycle. This may be indicative that KGD1 expression is conserved and related to genes involved in cellular growth. These results are also slighted since the represent evolution in the laboratory and not evolution in a constantly changing environment. (Figure was obtained by searching KGD1 at Expression connection.)
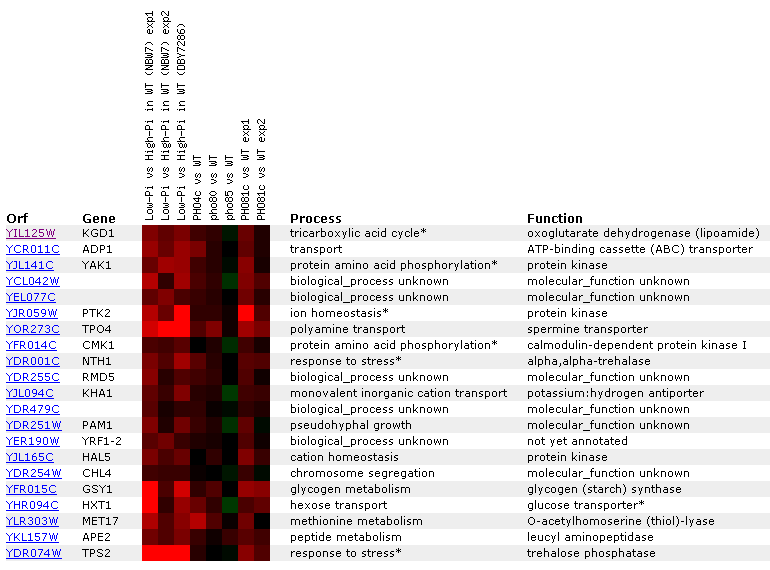
Figure 11: KDG1 expression regulated by the PHO pathway. KGD1 is moderately induced in the first three experiments where yeast grown with low phosphate concentrations are compared to yeast grown in high phosphate concentrations. KGD1 does not correlate well with other TCA cycle enzymes for this experiment, but we notice that its expression correlates with genes responding to stress.
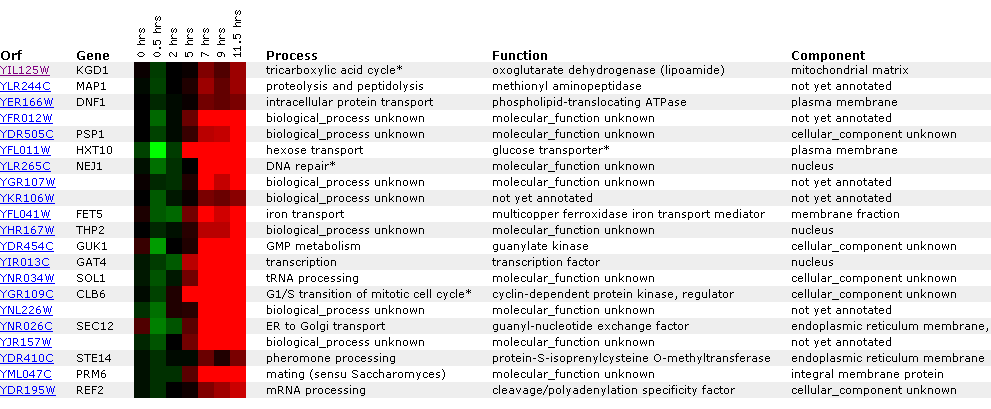 Figure
13: Graph representing KGD1 expression profile during sporulation. After initial
repression, KGD1 induction fluctuates between hours 5 and 10. Qualitatively,
KGD1 is induced after approximately 5 hours.
Figure
13: Graph representing KGD1 expression profile during sporulation. After initial
repression, KGD1 induction fluctuates between hours 5 and 10. Qualitatively,
KGD1 is induced after approximately 5 hours.
Conclusions: The data supports three conclusions. First, anytime yeast have to alter their metabolism they respond by changing the expression levels of the metabolic pathways by undergoing a stress response (see diauxic shift experiment). This suggests that cells must undergo major changes in order to alter metabolism. In the diauxic shift experiment, KGD1 correlated with TCA cycle genes, but the correlation coefficient was lower than .8. Second, KGD1 expression correlates well with genes involved in the cell cycle. The question is whether this is coincidence, or whether or not KGD1 is involved in the cell cycle. Third, the data does not support my hypothesis that genes in a similar chromosomal region are expressed similarly. This indicates that gene expression is highly regulated and very specific (not a big surprise). However since KGD1, KGD2 and LPD1 all encode subunits of the 2-oxogluterate dehydrogenase complex why are they not expressed similarly? Do they correlate below 0.8? Its time for another database search.......
YIL127C

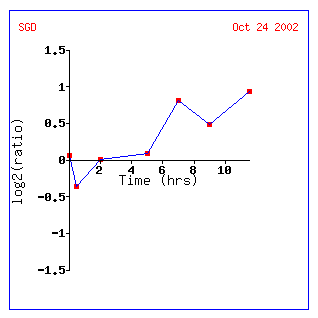
Figure 14: Expression for YIL127C for different alpha concentrations. The non-annotated gene is initially repressed. It is also correlated with genes involved in RNA processing and modification.

Figure 14: Expression profile for YIL127C after exposure to alpha factor. YIL127C is slightly repressed after 50 minutes (data not shown) and repressed after 90 minutes. YIL127C is being grouped with genes affecting t and r RNA processing and modifications.
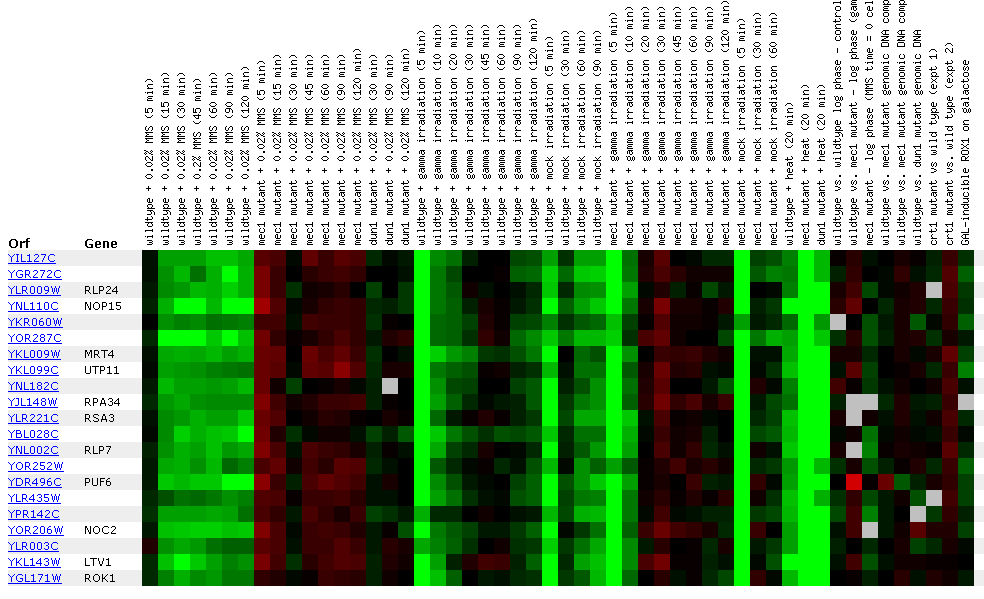 Figure
21: YIL127C
Expression for DNA damaging experiment. YIL127C is repressed during MMS
exposure and slightly induced in the mec1 mutant when exposed to 0.02% MMS
during the initial data points. Gamma light appears to repress YIL127C in
the initial data points as well as in mock irradiation. For the Mec1 mutants,
gamma irradiation represses then induces YIL127C during the first data points.
Also heat represses YIL127C in wild type, and mec1 and dun1 mutants.
Figure
21: YIL127C
Expression for DNA damaging experiment. YIL127C is repressed during MMS
exposure and slightly induced in the mec1 mutant when exposed to 0.02% MMS
during the initial data points. Gamma light appears to repress YIL127C in
the initial data points as well as in mock irradiation. For the Mec1 mutants,
gamma irradiation represses then induces YIL127C during the first data points.
Also heat represses YIL127C in wild type, and mec1 and dun1 mutants.
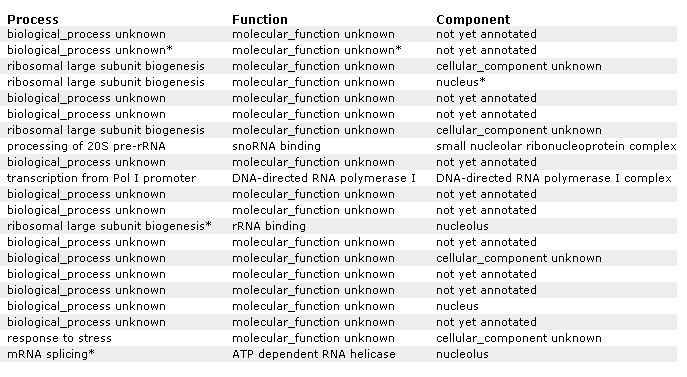
Figure 21 A: Supplemental information for figure 21. Designation of each gene and non annotated gene. Again, YIL127C is listed at the top and correlated with genes affecting RNA and ribosomes.
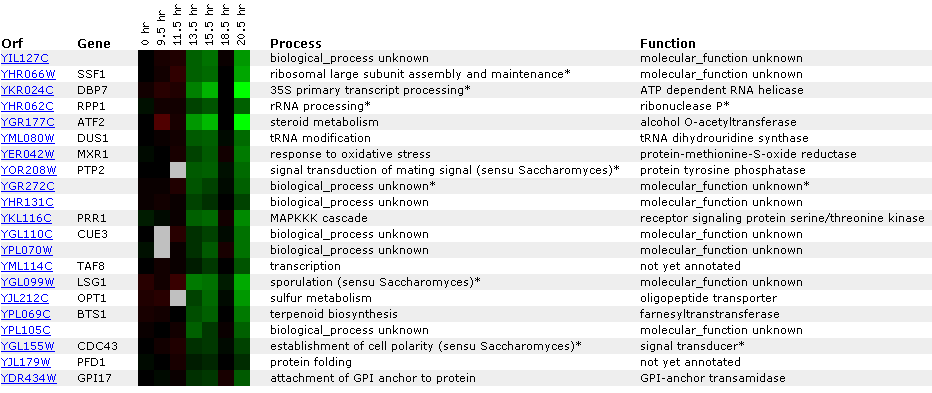

Figure 22: Experiment for diauxic shift. YIL127C is repressed at time points 4 and 5 and then returns to 1:1 expression in time point six. This is unusual because you would expect linear repression or induction over time. Ideally, you would like to see another experiment with shorter time segments. At time point seven, YIl127C is repressed again. Again YIl127C is correlated with ribosome, and RNA processing or modification genes. However, the SSF1 gene encodes a protein for 51,000+ kda (click here for results), thereby making it an unlikely match to YIL127C.
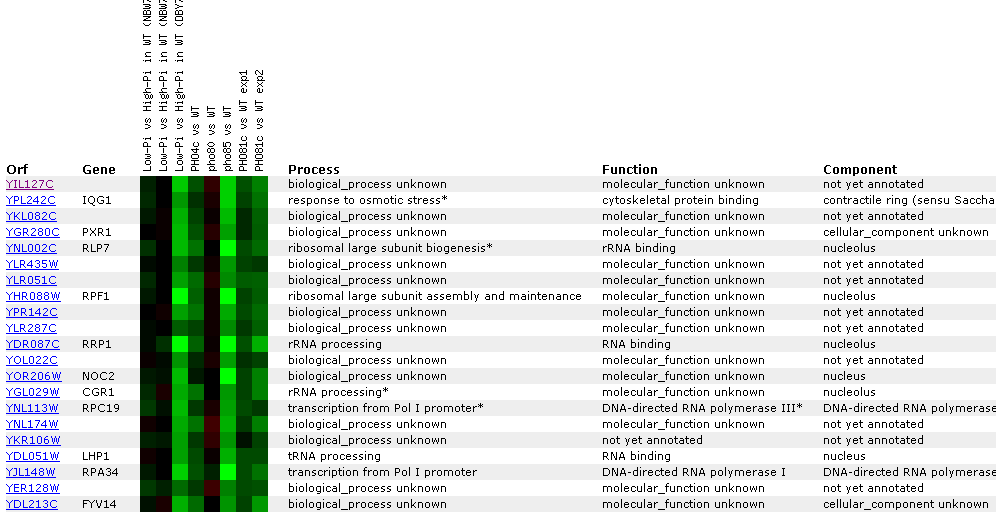 Figure
23: YIL127C
Expression regulated by the PHO pathway. For several different experiments
YIL127C is repressed. The key point in this figure is that YIl127C is correlated
with RRP1, RLP7, and CGR1- genes associated with either rRNA processing, or
rNA binding. YIL12C also correlates with tRNA genes.
Figure
23: YIL127C
Expression regulated by the PHO pathway. For several different experiments
YIL127C is repressed. The key point in this figure is that YIl127C is correlated
with RRP1, RLP7, and CGR1- genes associated with either rRNA processing, or
rNA binding. YIL12C also correlates with tRNA genes.
Figure 24: Environmental response for YIL127C. While this figure is extremely cumbersome, one can ascertain 10 simple trends. First, over time heat shock represses YIL127C. Second, if the heat shock temperature gradient is deceasing, YIL127C expression is induced. Therefore, YIL127C is heat sensitive and prefers lower temperatures. Third, hydrogen peroxide represses YIL127C for the first several time points. Fourth, heat shock in combination with sorbital results in initial repression. Fifth, exposure to 1.5 mM results in a repressed expression profile. Heat shock (29-33C) in combination with 1 M sorbitol results in initial repression. Sixth, 2.5 mM DDT results in initial induction and later repression. Seventh, during nitrogen depletion YIL127C repression decreases and has one time point in which the gene is induced. Eight, YPD stationary phase experiments resulted in the repression of YIL127C. Nine, the Steady state temperatures from 17-21C results in YIL127C induction. Ten, steady state experiments at 33-36C results in repression. The most interesting point about this figure is the strong correlation between the listed genes. Moreover, YIL127C correlates with two rRNA processing genes and one rRNA modification gene. There also several genes with functions relating to ribosome's and RNA processing. This suggests YIL127C may be involved with RNA binding. The majority of the genes listed in this figure are localized in the nucleolus.
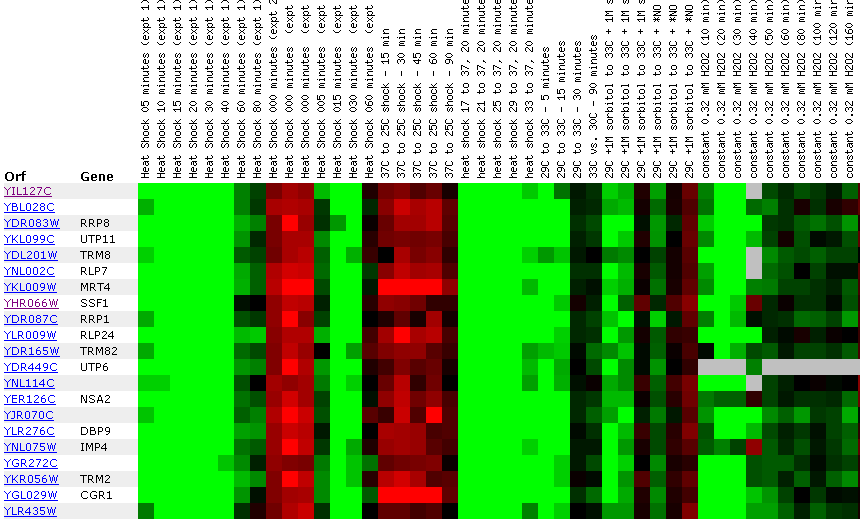
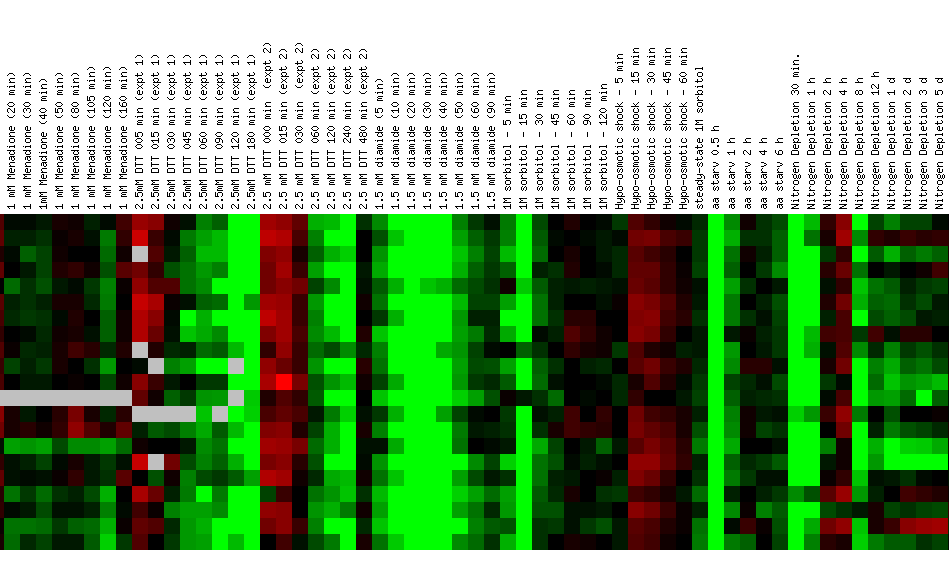
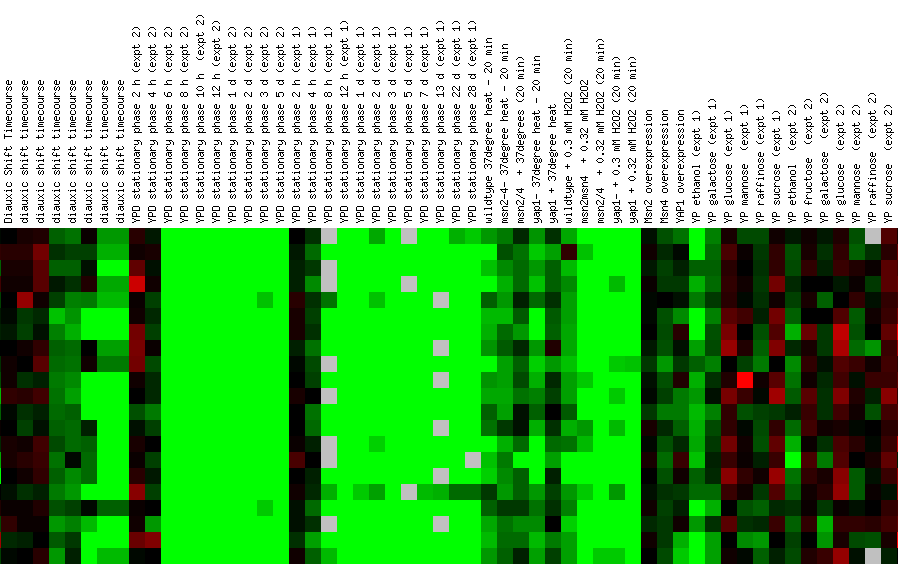
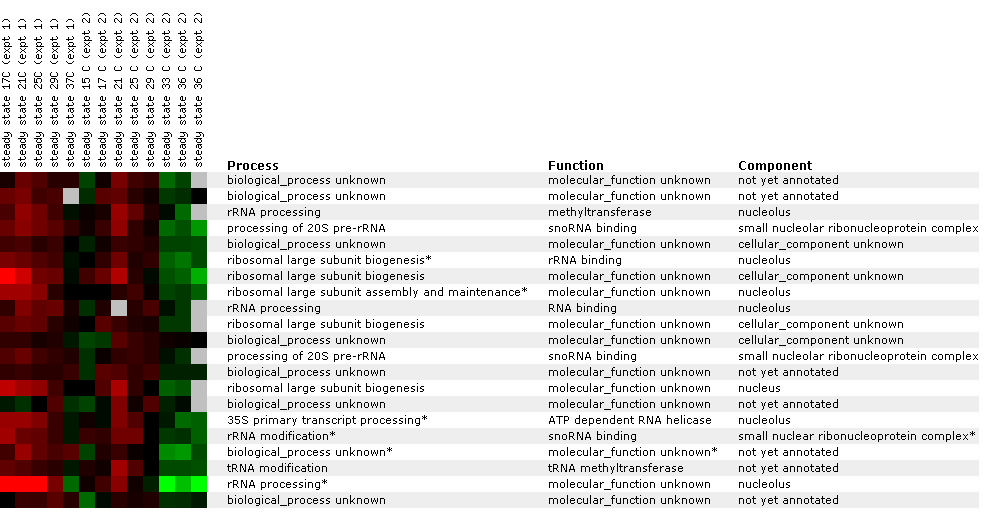
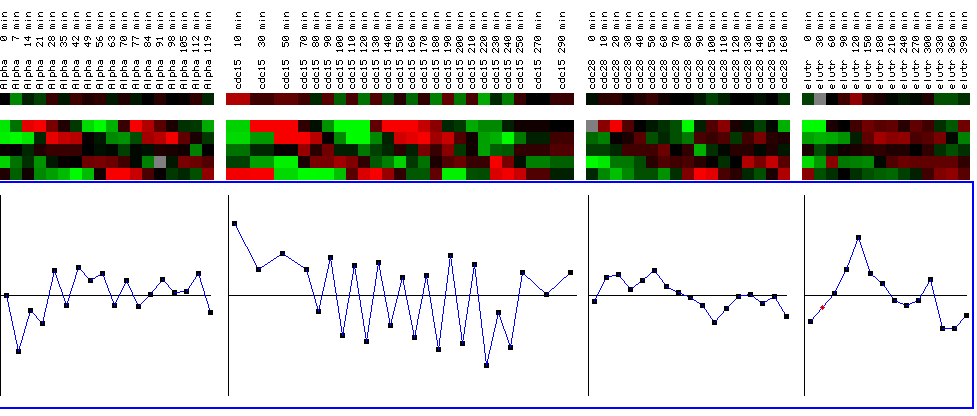
Figure 25: YIl127C expression profile for four different cell cycle experiments. The black bars represent a maximum and minimum log2 ratios of .5. The graphs represent YIL127C for the various time points for each experiment. The top line shows the expression profile for YIL127C, while the expression profiles below represent reference genes. While the expression profile does not alter change drastically in the first, third, and fourth experiments (going left to right), YIL127C is periodically repressed in the second experiment after being initially induced. While this is interesting, it doesn't yield any qualitative information concerning the function of YIL127C because the expression profile is not cyclic and therefore not representative of a gene involved in the cell cycle. In other words, YIL127C does not have an expression profile similar to the reference genes who's functions are related to the cell cycle. (Figure was obtained by searching KGD1 at Expression connection.)
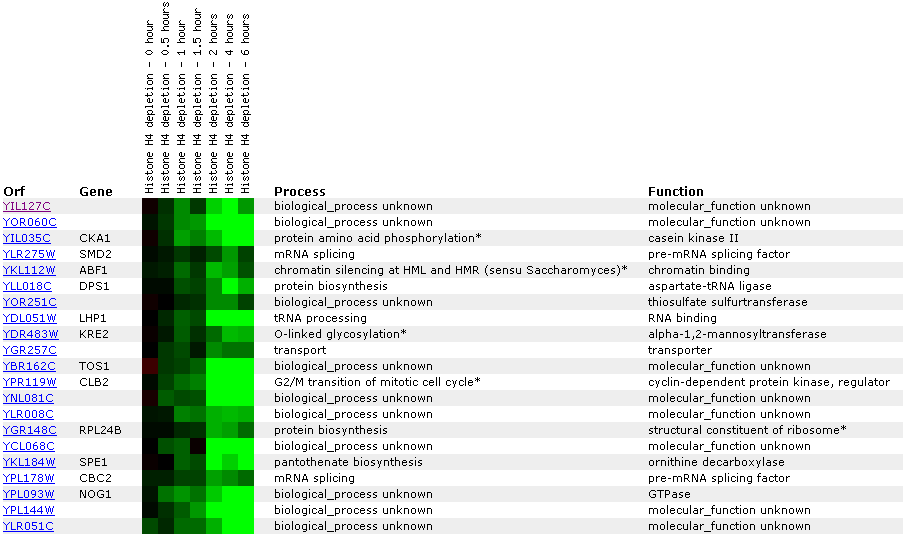 Figure
26: Expression
profile for histone depletion. YIL127C is primarily repressed during the
time course of histone H4 depletions. YIL127C appears to be correlated with
genes involved with RNA (LPH1, SMD2, CBC2).
Figure
26: Expression
profile for histone depletion. YIL127C is primarily repressed during the
time course of histone H4 depletions. YIL127C appears to be correlated with
genes involved with RNA (LPH1, SMD2, CBC2).
Conclusions: There are two major trends for YIL127C. First, the non-annotated gene expression correlates with genes with RNA functions. Specifically, genes involved in RNA processing, binding, and modification. However, there is one problem. Most of these genes encode for proteins with large molecular weights and YIL127C hypothetically encodes a protein with a small molecular weight. Second, YIL127C is temperature sensitive. In Figure 24, YIL127C is highly repressed at elevated temperatures.
DNA microarray data suggests my predictions from project 2 for YIL127C are incorrect. Using the guilt by association method, I would make the following predictions for YIL127C:
Biological Process: The data suggests YIL127C is responsible for regulation of RNA processing or modification
Molecular function: The data suggests YIL127C is probably involved in RNA binding. Since YIL127C encodes a small hypothetical protein, YIL127C could also be a transcription factor for genes involved in RNA processing and modification.
Cellular Componets: YIL127C would probably be localized in the nucleus or nucleolus along with most of the RNA processing and modification genes it correlated well with.
References:
1. Campbell, M., Heyer L. Dicovery Genomics Proteomics & Bioinformatics. Cold Spring Harbor Labratory and Benjamin Cummings. San Francisco. 2003.
2. Mockovciakova, D et al. The ogd1 and kgd1 mutants lacking
2-oxoglutarate dehydrogenase activity in yeast are allelic and can be differentiated
by the cloned amber suppressor. Current Genetics 24: 377-381 1993.
Davidson College Biology Main Page
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28035
Send comments, questions, and suggestions to: alcubre@davidson.edu