Malignant melanoma is a cancer of melanocytes in the skin. It is easily curable if detected early, but is very dangerous if it spreads throughout the body. It typically begins with a mole or dark place on the skin. It becomes dangerous when it spreads deeper into the skin and has access to other tissues. It is well documented that overexposure to sunlight can lead to malignant melanoma. It is also known that it can be an inherited trait (American Academy of Dermatology). Reading the Davies et. al. article introduces the idea that melanoma can be caused somatically by a mechanism other than UV light. Please read these links regarding melanoma to learn more about the disease.
Brief history of the relationship of BRAF and Melanoma.
Kyte-Doolittle Visualization of BRAF Sequence
Predator Secondary Structure Predictions

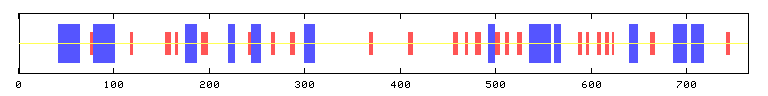
Alpha helix (Hh) : 165 is 21.57% (BLUE)
Extended strand (Ee) : 101 is 13.20% (RED)
Random coil (Cc) : 499 is 65.23% (YELLOW)
Figure 1: Secondary structure of BRAF based on PREDATOR computer model
This figure is taken from Network
Protein Sequence Analysis' Predator
Database
There are unfortunately no 3D images of BRAF or either of the RAF genes. I will provide, however, a picture and chime image of a MAP Kinase.
In order to understand the role that a mutation in BRAF plays in melanoma it is important to understand the RAS-RAF-MEK-ERK-MAP kinase pathway (to an extent). This pathway plays a role in cell differentiation, growth, and/or death. It is thus understandable that some of these genes could turn out to be oncogenes as is the case with BRAF. A great review article that deals with the genomic complexities of this pathway was written by Walter Kolch (this loads very slow, but its worth it). Of particular importance to the BRAF story is Figure 1 from this paper. From this figure we should observe the complexity of the pathway and the ways that phosphorylation plays a role in activating the pathway.
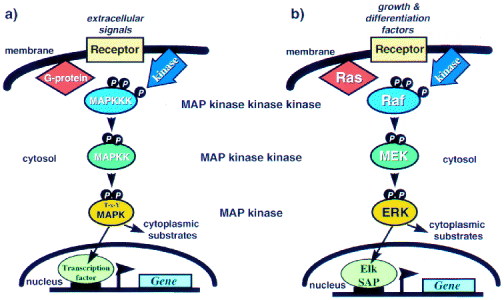
Figure 2 Schematic representation of the structure of MAPK pathways (a) General set-up of MAPK pathways; (b) the ERK pathway in particular.
This figure is available at http://www.biochemj.org/bj/351/0289/bj3510289.htm in a paper by Walter Kolch.
Information regarding this pathway's role in another cancer, Leukemia, is available in a paper by Lee and McCubrey.
Genetic Breakthrough in Skin Cancer Research. Cosmiverse. 10 June 2002. http://www.cosmiverse.com/news/science/science06100202.html
Maitland, Norman. Gene breakthrough will aid understanding of other cancers. The University of York Communications Office. 13 June 2002. http://www.york.ac.uk/admin/presspr/pressreleases/melanoma.htm
Meek, James. Gene project yields skin cancer breakthrough. Guardian Unlimited. 10 June 2002. http://www.guardian.co.uk/genes/article/0,2763,730577,00.html
The opening sentence of each of these sources creates a slightly different mood in the response of the reader. While it sound odd to use the term "mood" when referring to articles about cancer, we must remember that these stories are a means to sell either subscriptions or boost online advertisement referrals. The Guardian article magnifies the 'needle in a haystack' aspect of genetics research by referring to finding 1 mutation in 10 billion mutations that leads to malignant melanoma. The excitement is further enhanced by mentioning that this disease is "harsh" and kills " hundreds of British victims a year". The Cosmiverse article is slightly more conservative (and accurate) when it says that this mutation is involved in 66% of malignant skin cancers. This article also refers to the deadliness of the disease and states that 9,000 human die from the disease in the US and UK. The press release from The University of York does the most appropriate job of introducing readers to the reality of the breakthrough. They replace the phrase "the gene" with "a gene" when they describe what the researchers have discovered regarding a genetic cause of malignant melanoma. This article is similar to the others in creating mood by quoting a the statistic that this disease kills 1,600 people a year in the UK.
Another interesting issue mentioned in all of these articles is the relationship of UV rays from the sun and malignant melanoma. This was probably added as a way to make the reader involved in prevention of this disease. Readers can do nothing with the fact that single mutation causes BRAF to be "always on", nor can they take part in the development of a remedy for the disease (which they are told will take 5-15 years). By reminding readers of the link to sunlight exposure, the articles make readers leave the piece feeling that they have learned something they can introduce to their daily lives. While telling people to limit their UV exposure is a good thing, it is not necessarily in keeping with the facts of the BRAF story. If the authors of these three articles had read to the second page of the Davies et. al paper they would have seen that this mutation is not associated with UV light. UV light usually leads to pyrimidine dimers (thymine dimers for example); however, this mutation in BRAF is from a T to an A. The Guardian article says that 4 out of 5 cases of malignant melanoma are caused by exposure to sunlight, yet the researchers believe this mutation is somatically acquired in a different way.
All three articles discuss the effects this discovery could have on a treatment more than they discuss the discovery itself. They describe the mutation in BRAF as leading to BRAF being permanently turned on. The researchers they interview mention that all they have to do to fix the problem is to use a drug that will turn off the BRAF gene. The Guardian article mentions that Glivec is a drug that is already available that has a similar function. The important thing to take from this is that the articles make the situation seem extremely promising.
After reading these three popular press articles the situation stands as such:
Davies, H.; Bignell, G. R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague,
J.; Woffendin, H.; Garnett, M. J.; Bottomley,W.; Davis, N.; Dicks, E.; and 40
others. Mutations of the BRAF gene in human cancer. Nature 417: 949-954,
2002.
The authors of this paper began with an understanding of the role the RAS-RAF-MEK-ERK-MAP
kinase pathway plays in responding to growth signals. They also knew that in
15% of human cancer the RAS gene is mutated. RAF genes are serine/threonine
kinases that are regulated by RAS. The authors deduced that if mutation in RAS
can lead to cancer than mutations farther down the pathway could lead to the
same cancerous phenotypes. They thus began their study by examining genomic
DNA for the BRAF gene from 15 cancer cell lines and one healthy cell line (from
the same individual as the cancerous cell) to see if there were any variations.
They searched the BRAF exons and the intron/exon junctions for these differences.
They found 3 point mutations in the cancer lines that were not in the somatic
cells and thus concluded that these mutations occurred somatically. The authors
then examined the same sections of BRAF in 530 additional cancer cell lines.
43 of these cell lines had BRAF mutations and 20 of the 34 melanoma lines they
tested had mutations. This represented 59% of the melanoma cell lines tested,
and this was a much higher percentage than any of the other cancer types received
(see Figure 1 Davies et. al.). The authors realized that there was a trend of
BRAF mutations in cancer types that were known to be caused by RAS mutations.
This made BRAF an interesting lead to follow due to its direct relationship
with RAS.
The authors next discuss the single point mutation that is the focus of the
popular press articles describing this discovery. The mutation is at base 1796
and involves a T becoming an A. This is not in fact the only BRAF mutation that
leads to malignant melanoma, it is the mutation in 92% of the BRAF mutations
in melanoma. It is very interesting to point out that the authors state that
this point mutation is probably not caused by UV light because it is not a C
to T mutation as is usually seen when UV light is the culprit. They comment
that it is not surprising that melanoma is an outcome of the BRAF mutation because
in alpha melanocytes there is a cAMP pathway that upregulates cell division
through a pathway involving BRAF in response to UVB light. Supposedly other
cell types may not have a pathway that controls proliferation and differentiation
that relies so heavily on BRAF.
The authors found BRAF mutations in two places in the kinase domain. The mutation
at base 1796 leads to a amino acid substitution (V to E) at amino acid 599).
There were other mutations at 585, 594, 595, and 596. 89% of the BRAF mutations
were in the activation segment of the kinase domain (note how close the 5 mutated
amino acids are to each other). The authors point out that this portion of the
peptide is conserved all the RAF genes throughout evolution and thus we can
see why a mutation could be more problematic in this region (See Figure 2 in
Davies et. el. for BRAF homology map). The authors then discuss the second mutated
domain, the glycine-rich section of the kinase domain. These make up the other
11% of BRAF mutations in kinase domains. The authors chose to characterize the
effects of the V599E and V596Vand G464V and G468A mutations as they led to the
greatest number of melanomas.
When they tested the kinase activity of the different lines all had elevated
activity. V599E and V599E were very much higher than wildtype and V596V and
G464V were only slightly elevated. All four also showed an increased ability
to activate ERK in COS cells. As would be predicted, the two mutants with more
kinase activity showed greater phosphorylation of ERK. The researchers performed
a few more assays that I will not go into here regarding folding activity and
transformation ability of the mutant cells. They summarize these results by
saying that these mutations in BRAF activate the kinase. BRAF activity is regulated
by phosphorylation of its activation segment (the segment mutated in 92% of
malignant melanomas). Specifically in BRAF amino acids 598 and 601 must be phosphorylated
to activate kinase activity. The mutation at amino acid 599 (the most common
mutation) is said to mimic this phosphorylation because a negative amino acid
is put next to the sight of normal phosphorylation. Understanding on how the
other mutations lead to a mimicking of phosphorylation of BRAF is not yet understood.
They also point out that amino acid 599 is particularly important because mutations
at 600 are not found in cancers. The authors mention that the mutation in the
glycine-rich residue that leads to kinase activation may be an important insight
into other constitutively activated kinases.
Another important discovery discussed in the paper is which BRAF mutations
require RAS for kinase activation and which do not. It appears that only the
mutation at amino acid 599 does not require RAS in order to become active (due
to phosphorylation). Yet another cool finding in the paper involves a comparison
between BRAF and RAF1. The authors wonder why BRAF is a common cause of melanoma
and RAF1 is not. The answer appears to be that in RAF1 2 phosphorylations are
required to activate the kinase while in BRAF one of the two amino acids is
constitutively phosphorylated. Therefore we see that it takes fewer mistakes
to lead to an overly active BRAF versus RAF1.
The authors end the paper by referring to the possible treatments for melanoma
that their findings point to. It is put very briefly that inhibiting BRAF activity
may serve to treat this cancer. They make the point that the type of systematic
search they performed for somatic sequence variation will be very helpful in
the future of genomic research. This technique led them down a seemingly clear
path based on patterns of mutation locations. This paper is very important to
understand when learning Genomics because it discusses the complex issues of
protein regulation
Now that you have sailed through three popular press articles and stumbled through one scholarly article, you may be wondering how much reading is really necessary when it comes to new discoveries in Biology.
By comparing the scholarly and popular sources regarding this genetic breakthrough we have learned that the two sources do not aim for the same level of exploratory power. One difference is that in the popular articles facts and figures seem variable as evidenced by the different figures for number of people killed by melanoma each year and misstating statistics from the journal article. The primary difference is of course the level of complexity. The journal article, because it is very new, takes the genomic approach to the issue. It considers the extremely complex signaling pathway involved in what the popular articles simply call “turning the safety catches on”. By exposing the fact that not only are numerous genes/proteins involved in melanoma, but each protein can be activated to varying degrees of “on” or “off” based on differential phosphorylation, the journal article avoids the reductionism of the popular press articles. While it may seem that there is no need to expose the reader of online science articles to the genomic reality of the situation, it may be just as unnessecarry to keep them in the dark as to how complicated things really are. Its not that they need to have the RAS-RAF-MEK-ERK-MAP kinase pathway explained to them, rather they need to understand that there are many players even when one nucleotide leads to the mutation in many cases. This leads to another important difference in the two presentations, namely that there is no mention of the 4 other important mutations that lead to malignant melanoma in the popular press articles. The scientific authors performed extensive tests on how the differently located mutations lead to kinase activation. A mention of these tests would have shown that regulatory pathway based cancer isn’t really a matter of on/off. Finally, the popular articles focused primarily on the treatments that the finding would lead to. In the journal article there is only one sentence leading in this direction. It seems that the authors of the paper are quoted in the popular articles regarding the important impact this finding would have on treatment; however, since they didn’t mention this in their paper very extensively, it should be viewed only as a hypothesis.
Genetic Breakthrough in Skin Cancer Research. Cosmiverse. 10 June 2002. http://www.cosmiverse.com/news/science/science06100202.html
Maitland, Norman. Gene breakthrough will aid understanding of other cancers. The University of York Communications Office. 13 June 2002. http://www.york.ac.uk/admin/presspr/pressreleases/melanoma.htm
Meek, James. Gene project yields skin cancer breakthrough. Guardian Unlimited. 10 June 2002. http://www.guardian.co.uk/genes/article/0,2763,730577,00.html
Davies, H.; Bignell, G. R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M. J.; Bottomley,W.; Davis, N.; Dicks, E.; and 40 others. Mutations of the BRAF gene in human cancer. Nature 417: 949-954, 2002.
Kolch, Walter. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351: 289–305. 2000.
Lee JT Jr, McCubrey JA. The Raf/MEK/ERK signal transduction cascade as a target for chemotherapeutic intervention in leukemia. Leukemia 16(4):486-507. 2002.
Contact Kevin James with any comments or questions.
Visit Davidson Genomics