SER2 (also annotated as YGR208W) codes for a phosphoserine phosphotase involved in serine and glycine biosynthesis. It is found on chromosome VII, position 911888 to 912817 (SGD 2004;<http://db.yeastgenome.org/cgi-bin/locus.pl?locus=ser2>). An image of the open reading frame map of chromosome VII where SER2 is located can be found here. The genomic sequence can be found here. The coding sequence can be found here. The SER2 protein is predicted to have a transmembrane domain within its 309 amino acids (SGD 2004; <http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000003440>). SGD's list of compiled literature on SER2 can be found here.
Molecular Function: The SER2 protein is the only yeast protein involved in phosphoserine phosphotase activity (Melcher, Entian 1992, SGD 2004; <http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=4647>). In general, this type of molecule is a catalyst for the reaction of a L (or D) - O - phosphoserine and water molecule to be converted to the amino acid serine and a phosphate molecule (SGD 2004; <http://db.yeastgenome.org/cgi-bin/GO/go.pl?goid=4647>).
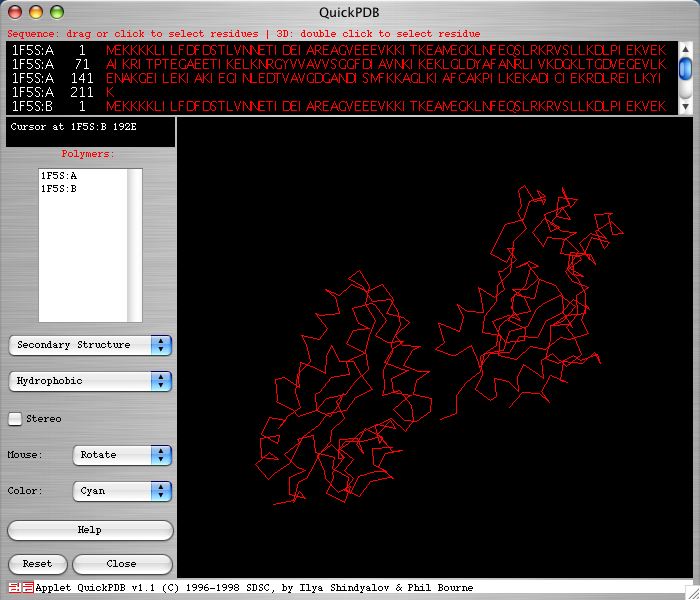
The SGD and PDB provide genes homologous to SER2 in other species. However, the level of homology is not very high, with the greatest similarity only registering a 40% identical and 32% similarity score (SGD 2004; <http://db.yeastgenome.org/cgi-bin/protein/get3d?sgdid=S000003440>). Below is an image of the two chains of Methanococcus jannaschii's 1f5s gene taken from the PDB web site. Notice that there are two separate chains in M. jannaschii's homologous protein. There is no literature available that suggests that the SER2 protein is cleaved to make two chains.

<http://www.rcsb.org/pdb/cgi/explore.cgi?job=graphics&pdbId=1F5S&page=>
Biological Process: The SER2 protein is one of several known to be involved in the synthesis of the amino acids serine and glycine. SER2 makes L-serine from 3 -phospho - serine, and L-serine is in turn made into L-glycine by two other proteins. Below is the SGD's illustration of the pathway:
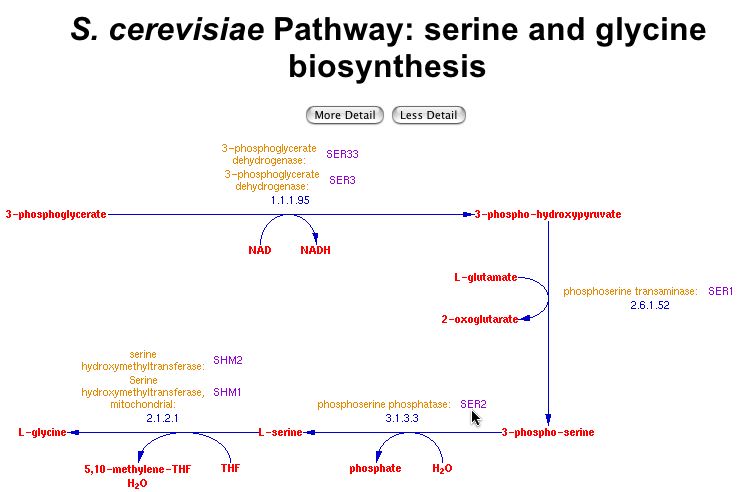
This image was reproduced with permission from SGD. <http://pathway.yeastgenome.org:8555/YEAST/new-image?type=PATHWAY&object=SER-GLYSYN-PWY&detail-level=2>
Mutants in SER2 show a variety of phenotypes depending on the mutations within the gene. Mutants are unable to produce serine, but other phenotypes include: sensitivity at five generations when grown in minimal +his/leu/ura medium; sensitivity at 20 generations when grown in 1M NaCl; sensitivity at 15 generations when grown in minimal +his/leu/ura medium; sensitivity at five generations when grown in synthetic complete -thr medium; apparent moderate growth defect on YPD after 20 and 60 generations; severe growth defect on minimal media and on lactate, and moderate growth defect when grown on NaCl (SGD 2004; <http://db.yeastgenome.org/cgi-bin/phenotype/phenotype.pl?phenotype=6402>). The mutants were made either by random insertion mutations or by whole gene deletions. (SGD 2004, SGD Curated Paper; <http://db.yeastgenome.org/cgi-bin/reference/reference.pl?dbid=S000074659> and Giaever et al. 2002).
Cellular Component: As stated in the overview, SER2 is found on chromosome VII. An image of the gene on the chromosome is located in the overview section.
Huh et al. inferred from a Direct Assay that SER2 is found both in the cytoplasm and the nucleus (2003; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=14562095>). As SER2 is involved with the production of amino acids, it is logical that the gene product would be found in the cytoplasm and not in other structures, such as the plasma membrane. However, a transmembrane domain has been predicted by SGD based on amino acid sequence analysis and predicted protein structure. The exact structure of SER2 is not known. The image below shows where in the protein the transmembrane domain is predicted to be:
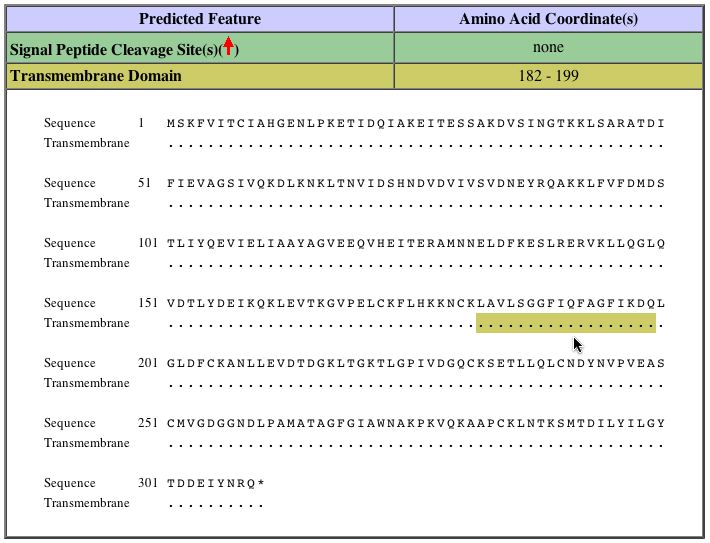
This image was reproduced with permission from SGD. <http://db.yeastgenome.org/cgi-bin/protein/protein?sgdid=S000003440>
The ORF YGR210C has not yet been annotated (SGD 2004; <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=ygr210c>). The function, structure, and biological process is yet unknown. YGR210C is located on chromosome VII, two genes from SER2 and on the opposite strand. The image below points out YGR210C, with SER2 to the left.
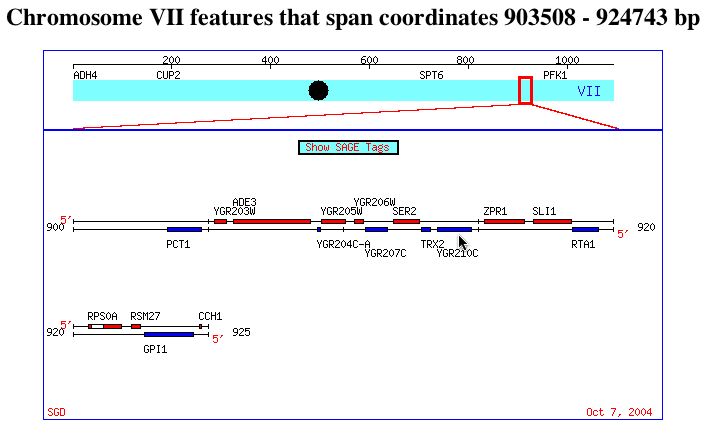
This image was reproduced with permission from SGD. <http://db.yeastgenome.org/cgi-bin/ORFMAP/ORFmap?dbid=S000003442>
SGD offers that YGR210C is found in the cytoplasm (Huh et al. 2003; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=14562095>) and there are some mutant phenotypes known. In systematic deletion of the gene, the mutant strain was found to be viable, but during genetic footprinting, phenotypes such as a moderate growth defect after 20 and 60 generations when grown on YPD, and moderate growth defects when grown (separately) on minimal media, lactate, and NaCl (Giaever et al. 2002; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=12140549> and SGD 2004, SGD Curated Paper; <http://db.yeastgenome.org/cgi-bin/reference/reference.pl?dbid=S000074659>). Further, a study by Guerreiro and Rodrigues-Pousada revealed that YRG210C is not an essential gene to the functioning of yeast cells (2001; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=11427960>).
A BLASTn search of the genomic sequence (found here) came back with these results: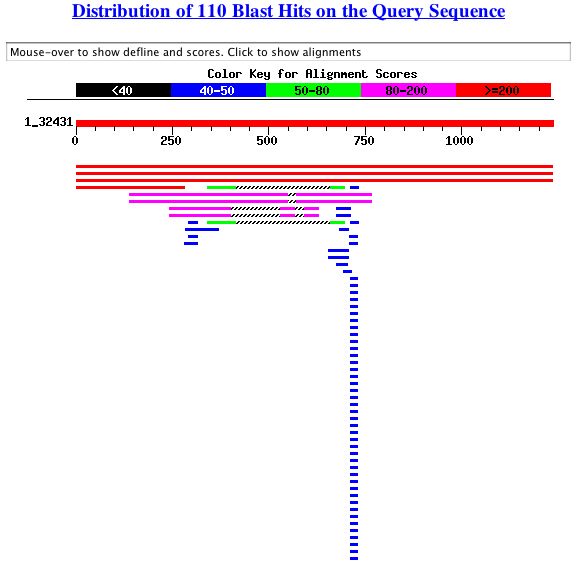
The most significant results are shown below:
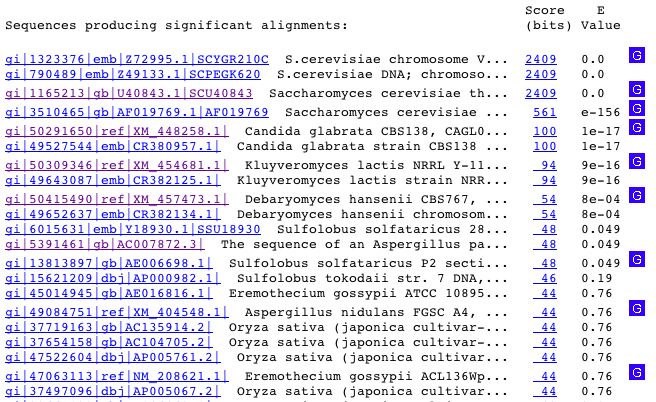
The first two results are from the ORF YGR210C. The third results in a match to the S. cerevisiae thioredoxin-2 (TRX2) gene (<http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=Nucleotide&list_uids=01165213&dopt=GenBank>). Conveniently, TRX2 is adjacent to YGR210C. This produces a discrepancy between the two databases. SGD separates TRX2 and YGR210C while the BLAST search and the NCBI database recognize YGR210C as a part of TRX2 at 99% identity match. The SGD database was most recently updated in 2004, compared to the 1995 entry of TRX2 (which went unpublished as well). For clarity's sake, it will be assumed that the SGD database is correct and that TRX2 and YGR210C are in fact two separate ORFs.
The fourth result in BLAST is the most promising. The sequence input matches 100% to the ZPR1 gene which encodes for a zinc finger protein. The match is shown below:
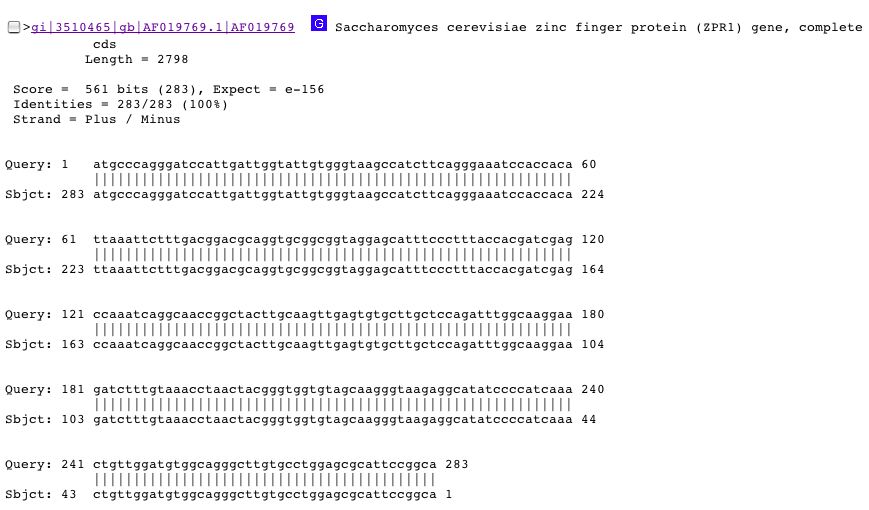
ZPR1 is a zinc finger protein that is found in the nucleolus of proliferating cells, but in the cytoplasm at all other times. Cytoplasmic location of ZPR1 corresponds to the location of YGR210C. As of now, there is no known function for nuclear ZPR1. Mutations in the zinc finger gene render the cells not viable and establish this gene as an essential gene, a completely different phenotype and classification of mutants of YGR210C (Galcheva-Gargova et al. 1998; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed&cmd=Retrieve&list_uids=9763455&dopt=Citation>).
Several of the other hits from the BLAST search show that YGR21C is similar to sequences in Candida glabrata strain CBS138, Kluyveromyces lactis NRRL Y-1140, and Debaryomyces hansenii CBS767. The sequences in the other species have no known function, and are merely predicted mRNAs. These similarities were found by Dujon et al. in a study of the evolution of yeast species (2004; <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed&cmd=Retrieve&list_uids=15229592&dopt=Citation>).
These findings have several implications. First, YGR210C could be a homologue to ZPR1 because of their highly similar genomic sequences and common location in the cell. However, the difference in ZPR1 being an essential gene and YGR210C not being essential makes the argument that they have different functions. However, if these two ORFs are homologues, then there could be a mutation in a genomic sequence that controls the expression of YGR210C rendering YGR210C a nonfunctional gene. For example, a mutation in a promoter sequence would cause YGR210C not to be expressed properly. The second implication is that YGR210C could be an orthologue to the similar genomic sequences found in the three other species mentioned above. As an orthologue, YGR210C would be under selective pressures to maintain the conservation of the sequence between the species of yeast, implying that there is an important, but not necessarily essential functioning gene product.
From these findings of YGR210C's genomic sequence, I propose that the ORF codes for a zinc finger protein, similar or homologous to ZPR1 protein, and that YGR210C is othologous to the similar sequences found in the other yeast species.
Dujon B, et al. 2004 Jul. Genome evolution in yeasts [abstract]. In Nature 1;430(6995):35-44. PubMed Database <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed&cmd=Retrieve&list_uids=15229592&dopt=Citation>. Accessed 2004 Oct 8.
Galcheva-Gargova Z, et al. 1998 Oct. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells [abstract]. In Mol Biol Cell. 9(10):2963-71. PubMed Database <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed&cmd=Retrieve&list_uids=9763455&dopt=Citation>. Accessed 2004 Oct 8.
Giaever G, et al. 2002 Jul. Functional profiling of the Saccharomyces cerevisiae genome [abstract]. In Nature 25;418(6896):387-91. PubMed Database <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=12140549>. Accessed 2004 Oct 7.
Guerreiro P and Rodrigues-Pousada C. 2001 Jun. Disruption and phenotypic analysis of six open reading frames from chromosome VII of Saccharomyces cerevisiae reveals one essential gene [abstract]. In Yeast 30;18(9):781-7. PubMed Database <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=11427960>. Accessed 2004 Oct 7.
Huh WK, et al. 2003 Oct. Global analysis of protein localization in budding yeast [abstract]. In Nature 16;425(6959):686-91. PubMed Database <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=14562095>. Accessed 2004 Oct 7.
Melcher, K and Entian KD. 1992 Apr. Genetic analysis of serine biosynthesis and glucose repression in yeast [abstract]. In Curr Genet. 21(4-5):295-300. PubMed Database <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=1326413>. Accessed 2004 Oct 7.
Saccharomyces Genome Database [SGD]. 2004 Sep 20. SER2/YGR208W. <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=ser2>. Accessed 2004 Oct 5.
Saccharomyces Genome Database [SGD]. 2004 Sep 20. YGR210C. <http://db.yeastgenome.org/cgi-bin/locus.pl?locus=ygr210c>. Accessed 2004 Oct 5.
Saccharomyces Genome Database [SGD]. 2004. SGD Curated Paper: Genetic footprinting: A functional analysis of the S. cerevisiae genome. <http://db.yeastgenome.org/cgi-bin/reference/reference.pl?dbid=S000074659>. Accessed 2004 Oct 7.
Contact Megan at mecastle@davidson.edu