My Favorite Yeast Genes
This web page was produced as an assignment for an undergraduate course at Davidson College.
| My favorite annotated yeast gene: LAS 17/BEE1 | ||||||||||||||
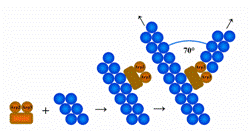
LAS17, also known as BEE1, creates a protein known as an actin assembly factor in Saccharomyces cerevisiea. More specifically, this gene codes for the protein that regulates Arp2/3 (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707). This protein is responsible for allowing new actin filaments to grow from a "mother" actin filament. They bind to the "mother" filament and allow the new actin filament to polymerize at a 70 degree angle from the "mother" filament (Wikipedia "Arp 2/3 Complex", http://en.wikipedia.org/wiki/Arp2/3_complex). LAS17 in yeast is a known homolog to the Wiskott-Aldrich Syndrome protein in humans (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707). |
||||||||||||||
| Actin in Yeast | ||||||||||||||
Actin structure, Permission pending from Wikipedia.com |
|
|||||||||||||
| Location of LAS17/BEE1 | ||||||||||||||
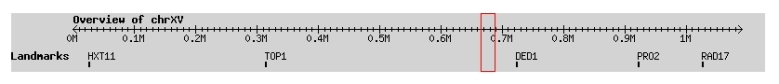
LAS17/BEE1 is located on chromosome 15 at positions 665,940 - 687,841 bases. This chromosome is the third largest in the yeast genome coding for 9% of the genome. This chromosome contains 92 other genes in addition to LAS17/BEE1 (Dujon et al. 1997, http://www.ncbi.nlm.nih.gov/entrez/ |
Figure 1. Location of genes found below on chromosome 15. (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707) |
|||||||||||||
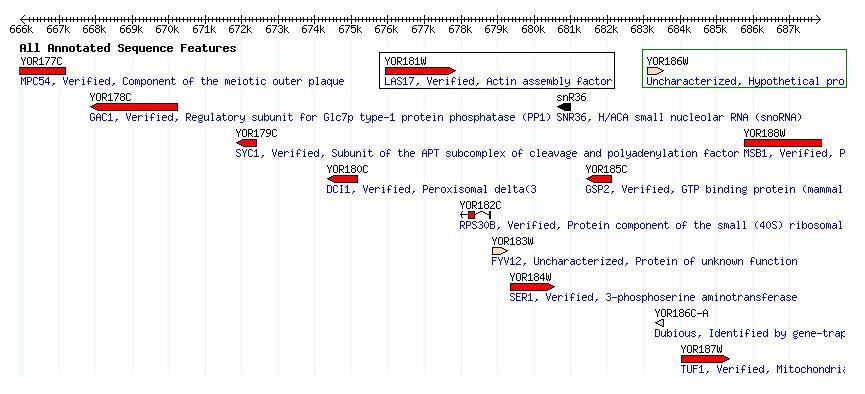
Figure 2. Genes and open reading frames located close to LAS17/BEE1. The systematic name of this gene is YOR181W. This gene is enclosed in a box above. (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707) |
||||||||||||||
| DNA Sequence of LAS17: | ||||||||||||||
| ATGGGACTCCTAAACTCTTCAGATAAGGAAATTATCAAAAGGGCTCTACCAAAAGCGTCG AATAAGATTATTGATGTTACGGTGGCTCGACTATACATTGCATACCCTGATAAAAATGAA TGGCAGTACACTGGACTTTCAGGAGCTCTTGCTCTAGTAGACGATCTTGTGGGGAATACT TTTTTTTTGAAATTAGTTGACATCAATGGCCATAGAGGAGTTATCTGGGACCAAGAATTG TATGTGAATTTTGAATACTATCAAGACCGTACTTTTTTTCATACATTTGAGATGGAAGAA TGCTTTGCAGGTTTATTGTTTGTAGATATTAATGAAGCATCGCACTTTTTAAAGAGAGTT CAAAAGCGTGAAAGATATGCTAACAGGAAAACTTTGTTGAACAAAAATGCTGTAGCATTA ACCAAGAAAGTAAGAGAAGAACAAAAATCTCAAGTGGTGCACGGCCCAAGAGGGGAGTCA TTGATTGACAATCAAAGGAAAAGATATAATTATGAAGATGTGGACACAATTCCAACTACA AAGCATAAGGCTCCTCCCCCTCCTCCGCCAACGGCCGAAACATTTGATTCAGACCAAACA AGTTCATTTTCCGATATCAATTCGACAACAGCATCCGCACCGACTACCCCAGCCCCTGCT CTTCCTCCTGCATCTCCTGAAGTAAGAAAAGAAGAAACGCATCCAAAGCATAGTTTACCG CCTTTACCAAATCAGTTTGCGCCATTACCAGACCCTCCACAACATAACTCTCCACCTCAA AATAACGCGCCTTCGCAACCCCAAAGCAATCCATTTCCATTCCCAATTCCTGAAATTCCC TCGACACAGTCTGCAACAAACCCATTTCCATTTCCGGTACCTCAGCAGCAGTTTAATCAA GCTCCTTCAATGGGCATACCACAGCAGAATAGGCCCCTTCCACAGTTGCCTAACAGAAAT AATCGGCCTGTGCCACCTCCTCCGCCAATGCGTACCACTACTGAAGGTTCAGGTGTTCGC CTACCTGCTCCTCCACCTCCGCCAAGGCGTGGGCCAGCACCACCGCCTCCACCACATAGG CACGTAACCAGTAATACCCTGAATTCTGCCGGTGGAAATAGCCTCCTTCCACAGGCCACT GGAAGAAGAGGGCCAGCACCACCACCTCCTCCAAGAGCATCTCGCCCCACACCAAACGTT ACGATGCAACAAAATCCACAACAGTACAATAATTCTAACCGCCCCTTTGGATATCAGACA AATAGCAACATGTCATCTCCACCCCCTCCTCCAGTGACAACTTTCAATACCCTGACACCA CAAATGACTGCAGCAACTGGACAACCTGCAGTTCCCCTTCCTCAGAATACTCAAGCACCT TCGCAAGCCACAAATGTGCCAGTGGCACCACCACCTCCTCCGGCATCTTTAGGCCAGTCG CAGATACCTCAGTCAGCACCCTCAGCACCTATTCCGCCAACGTTACCATCGACGACGAGT GCTGCACCACCTCCGCCACCAGCATTCCTAACTCAACAACCTCAATCTGGAGGAGCTCCA GCTCCACCCCCACCTCCTCAAATGCCAGCTACATCAACATCCGGAGGCGGTTCATTCGCT GAAACTACTGGAGATGCAGGTCGTGATGCACTTTTAGCTTCAATTAGAGGGGCAGGTGGC ATAGGCGCTTTGAGAAAAGTTGACAAATCGCAGCTAGATAAGCCCTCAGTTTTACTGCAG GAAGCACGTGGAGAATCTGCTTCACCACCAGCAGCGGCTGGAAATGGAGGCACACCTGGT GGACCTCCGGCTTCTTTAGCAGATGCGTTGGCAGCAGCTTTAAACAAAAGAAAAACTAAA GTGGGAGCTCATGACGATATGGACAATGGTGATGATTGGTAA (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707) | ||||||||||||||
| Amino Acid Sequence of LAS17: | ||||||||||||||
N-Terminal Sequence: MGLLNSS C-Terminal Sequence: MDNGDDW Length: 633 Amino Acids Molecular Weight: 67,571 Daltons (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707) |
||||||||||||||
| MGLLNSSDKE IIKRALPKAS NKIIDVTVAR LYIAYPDKNE WQYTGLSGALALVDDLVGNT FFLKLVDING HRGVIWDQEL YVNFEYYQDR TFFHTFEMEECFAGLLFVDI NEASHFLKRV QKRERYANRK TLLNKNAVAL TKKVREEQKSQVVHGPRGES LIDNQRKRYN YEDVDTIPTT KHKAPPPPPP TAETFDSDQTSSFSDINSTT ASAPTTPAPA LPPASPEVRK EETHPKHSLP PLPNQFAPLPDPPQHNSPPQ NNAPSQPQSN PFPFPIPEIP STQSATNPFP FPVPQQQFNQAPSMGIPQQN RPLPQLPNRN NRPVPPPPPM RTTTEGSGVR LPAPPPPPRRGPAPPPPPHR HVTSNTLNSA GGNSLLPQAT GRRGPAPPPP PRASRPTPNVTMQQNPQQYN NSNRPFGYQT NSNMSSPPPP PVTTFNTLTP QMTAATGQPAVPLPQNTQAP SQATNVPVAP PPPPASLGQS QIPQSAPSAP IPPTLPSTTSAAPPPPPAFL TQQPQSGGAP APPPPPQMPA TSTSGGGSFA ETTGDAGRDALLASIRGAGG IGALRKVDKS QLDKPSVLLQ EARGESASPP AAAGNGGTPGGPPASLADAL AAALNKRKTK VGAHDDMDNG DDW (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707) | ||||||||||||||
|
||||||||||||||
| Mutations: | ||||||||||||||
A null mutation is viable, but causes impaired cellular budding and cytokinesis, disrupted cortical actin, and some mutants may accumulate in secretory vessicles. If LAS17 becomes non-functional, S. cerevisiae is known to lose the ability to endocytose. Below are listed some protein interaction mutations that may occur if LAS17 becomes mutated. Dosage Rescue: An overexpression of LAS17 may compensate for a lethality or growth defect in another regulatory protein for actin reproduction. Phenotypic Enhancement: An overexpression of LAS17 may cause an overexpression of another protein. Phenotypic Suppression: An overexpression of LAS17 may cause an underexpression of another protein. Synthetic Lethality: Mutations of LAS17 and another gene related to actin may be lethal. Finally, the human homolog of LAS17 is associated with the Wiskott Aldrich Syndrome. (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707) |
||||||||||||||
| Wiskott Aldrich Syndrome | ||||||||||||||
| This syndrome is associated with a loss of actin function in leukocytes. The disease is fairly rare with only 4 cases in 1,000,000 individuals, but this disease is an X-linked recessive disorder affecting only males. This syndrome is associated with an immunoglobulin M deficiency that causes persistent thrombocytopenia and small platelets. Researchers assume these phenotypes are expressed because the mutation causes incorrect signalling and growth of hematopoietic cells. Males who express this disease are also at higher risks of developing autoimmune disease and hematologic malignancies. (Dibbern, http://emedicine.com/med/topic1162.htm) | ||||||||||||||
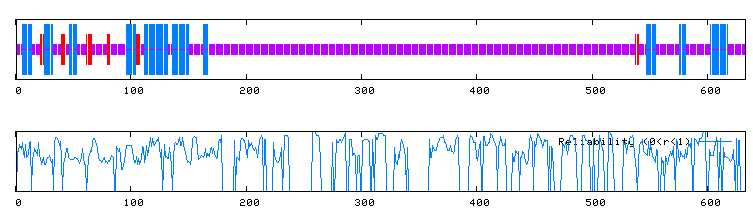
| Secondary Structure of LAS17 by Predator Analysis: | ||||||||||||||
Figure 3. a. Predator analysis of LAS17. A blue region indicates an alpha helix and red regions indicate extended strands. b. Reliability of Predator predictions. (Predator, http://npsa-pbil.ibcp.fr/cgi-bin/secpred_preda.pl) |
||||||||||||||
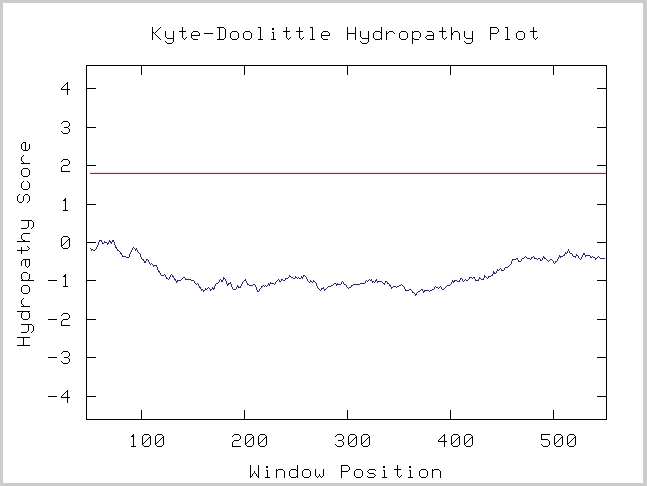
| Kyte-Doolittle Graph of LAS17: | ||||||||||||||
Figure 4. This figure shows that LAS17 does not have any transmembrane component, which confirms our predictions that LAS17 protein acts within the cytoplasm ((Kyte-Doolittle Hydropathy Plot, http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm). |
||||||||||||||
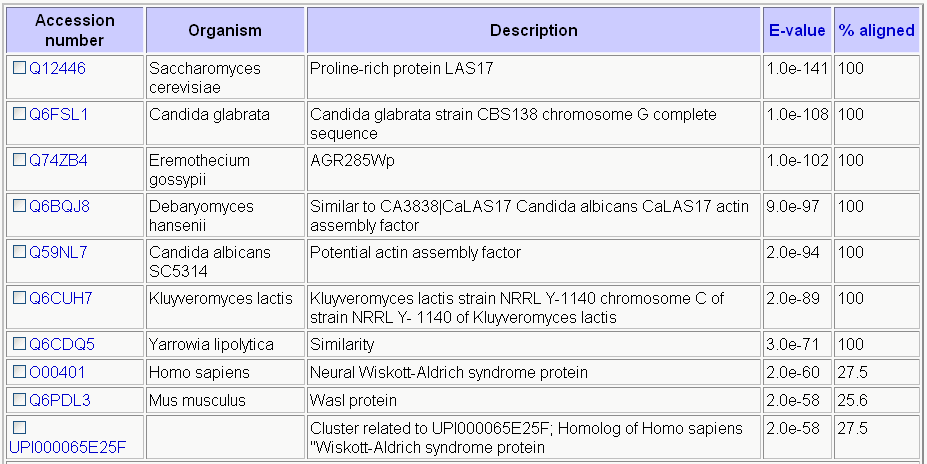
| Homologous Genes to LAS17: | ||||||||||||||
Table 1. Most homologous genes are found within other fungi, but homologs of LAS17 may be found in humans and mice (SGD, http://db.yeastgenome.org/cgi-bin/homolog/uniprotHomolog). |
||||||||||||||
| Blastp Results for LAS17: | ||||||||||||||
Table 2. The top results for LAS17 in a Blastp search. Most hits are hypothetical proteins in yeast until you reach other WASP proteins in mice and humans (Blastn, http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). |
||||||||||||||
Unannotated Yeast Gene: YOR186W |
||||||||||||||
| YOR186W has no known molecular function, biological function, or cellular components. | ||||||||||||||
Figure 5. Unannotated gene YOR186W is in the green box. This is one of the closest uncharacterized open reading frame to my favorite yeast gene LAS17 in the black box (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005712). |
||||||||||||||
DNA Sequence of YOR186W |
||||||||||||||
| ATGACTTTGGCTTTTACGACCTTCGCTATTTCCAAGATAAACAATTCGTCCACGAATGAA GATTCTAAAGTCATGATACTGTGCGATGAGCATCACCCTTTCGAGAAGGGTTACTTTAAG TCCGCCATACGAGCCTTCGGCAACAGCATTAAGCTAGGATTAATGGGTAACTCGCGACCA GAAGACGCAGCGTCCATATTCCAAGATAAAAATATTCCTCACGATCTGACGACGGAAGAA TTCAGGTTGCAGCTGGTCTGTATGGCTTTTTCCTGGTTCATATTTGGTCTCTTCATCGCC TGCTTACTACTTTGTATCACGCTTGTGCTAACATCGCGATATCCTGGAGAAAATGAAAAC AAAGCCACGGAAGTTGTGCCATCCAGTAATATTGACGACGAAGAAAAGCAATTATCGCTT TCCGACATGATTTAG (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005712) | ||||||||||||||
| Amino Acid Sequence of YOR186W | ||||||||||||||
N terminal: MTLAFTT C terminal: LSLSDMI Length: 144 Amino Acids Molecular Weight: 16,179 Daltons (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005712) |
||||||||||||||
| MTLAFTTFAISKINNSSTNEDSKVMILCDEHHPFEKGYFKSAIRAFGNSI KLGLMGNSRPEDAASIFQDKNIPHDLTTEEFRLQLVCMAFSWFIFGLFIA CLLLCITLVLTSRYPGENENKATEVVPSSNIDDEEKQLSLSDMI (SGD, http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005712) | ||||||||||||||
| Blastn Results for YOR186W | ||||||||||||||
Table 3. Few significant hits result from searching for the nucleotide sequence of the unannotated gene. |
||||||||||||||
Blastp Results for YOR186W |
||||||||||||||
Table 4. Few significant results exist for the amino acid sequence of YOR186W.
|
||||||||||||||
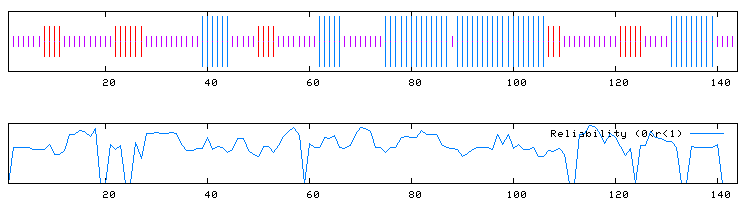
| Predator Secondary Structure Analysis of YOR186W | ||||||||||||||
Figure 6. a. Predator analysis of YOR186W. A blue region indicates an alpha helix and red regions indicate extended strands. b. Reliability of Predator predictions. (Predator, http://npsa-pbil.ibcp.fr/cgi-bin/secpred_preda.pl) |
||||||||||||||
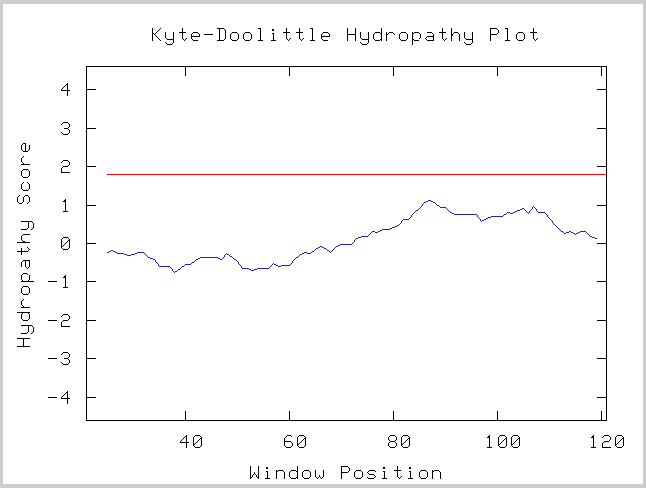
Kyte-Doolittle Hydropathy Plot |
||||||||||||||
Figure 7. This Kyte-Doolittle analysis shows that this open reading frame is likely to lack a transmembrane function. (Kyte-Doolittle Hydropathy Plot, http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm) |
||||||||||||||
|
||||||||||||||
| FLiP | ||||||||||||||
| Yeast cells frequently contain plasmids in addition to their genome. One such plasmid is a 2u circle that is 6.3 kilobases long that does not appear to confer an advantage to cells that carry the plasmid. This plasmid contains a gene called FliP that promotes duplication of the plasmid by inverting the sequence of the plasmid. This protein encourages recombination events within this plasmid that allows several copies of the plasmid to be produced during each genome replication event. (Maloy, http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/plasmids/yeast-plasmid.html) | ||||||||||||||
Proposed function of YOR186W |
||||||||||||||
| According to my research, I believe that YOR186W may be a non-functional open reading frame within the Saccharomyces cerevisiae genome that has been acquired through recombination between the 2u circle plasmid and chromosome 15. If this gene is functional, I suggest that it may assist with recombination between chromosomes or assist with protein binding to the DNA sequence. | ||||||||||||||
| Citations | ||||||||||||||
"Actin". Wikipedia. 22 Sept. 2006. Accessed 4 Oct. 2006. <http://en.wikipedia.org/wiki/Actin> "Arp2/3 Complex". Wikipedia. 18 July 2006. Accessed 4 Oct. 2006. <http://en.wikipedia.org/wiki/Arp2/3_complex> BlastN. NCBI. 2006. Accessed 4 Oct. 2006. <http://www.ncbi.nlm.nih.gov/blast/Blast.cgi> BlastP. NCBI. 2006. Accessed 4 Oct. 2006. <http://www.ncbi.nlm.nih.gov/blast/Blast.cgi> Dibbern, D.A. "Waskott-Aldrich Syndrome". E-Medicine. 15 June 2005. Accessed 4 Oct. 2006. <http://emedicine.com/med/topic1162.htm> Dujon, B. et al. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XV. Nature 387: 98 - 102. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=pubmed> Giaever, G. et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387 - 391. Maloy, S. "Plasmids in Eukaryotic Microbes". Accessed 4 Oct. 2006. <http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/plasmids/yeast-plasmid.html> Saccharomyces Genome Database. 2006. Accessed 4 Oct. 2006. <http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005707> Saccharomyces Genome Database. 2006. Accessed 4 Oct. 2006. <http://db.yeastgenome.org/cgi-bin/singlepageformat?sgdid=S000005712> Tools Used on This Website Kyte-Doolittle Hydropathy Plot. < http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm> Predator. < http://npsa-pbil.ibcp.fr/cgi-bin/secpred_preda.pl> |
||||||||||||||
Questions? Please contact Krcecala@davidson.edu
© Copyright 2006 Department of Biology, Davidson College, Davidson, NC 28035