This page was produced as an assignment for an undergraduate course at Davidson College.
How to do a Nucleotide BLAST
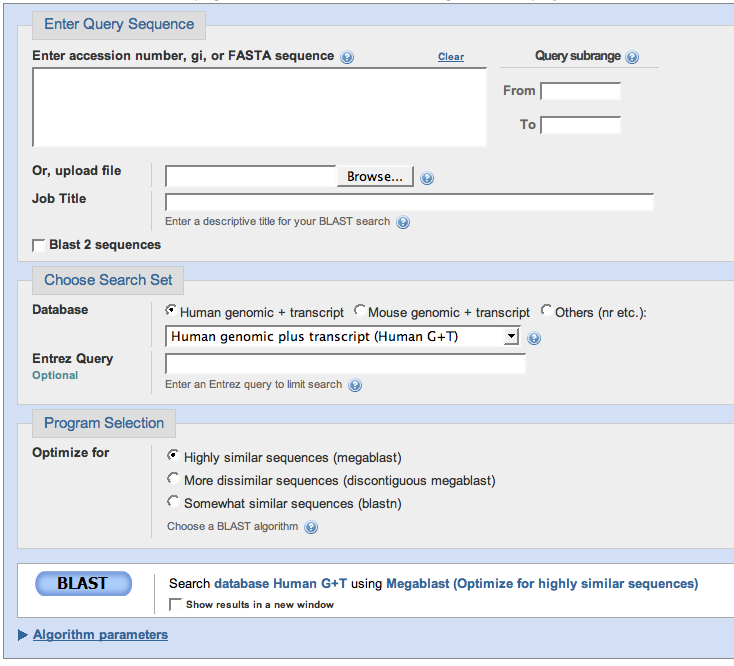
You have a nucleotide sequence and you want to compare it to other nucleotide sequences in the database! If you clicked the link above, you should arrive at a page that looks like this:

|
In the box "Enter Query Sequence" you should submit a nucleotide sequence. The one pictured on the right numbers the nucleotide sequence, but you can also enter a nucleotide sequence with no numbers in it or a FASTA sequence (a sequence where the first line starts with a ">" sign). You can also upload a text file with your sequence in it. |
|
|
In the box "Choose Search Set" you should select the database you want to search for your entered nucleotide sequence. If you know it came from the human or mouse genome, those options are readily available for you. If you're not sure where it came from, like my sequence, you should choose the "Nucleotide Collection (nr/nt)" option, which will search for your sequence in all nonredudant nucleotide databases. If you know it came from a species other than mouse or human, choose the "Nucleotide collection" option, then on the line underneath the selection box, type the name of the species you want to search. There are many other options on the pull-down menu to explore, but the most likely databases to be of use to you are the ones previously discussed. |
|
|
In the "Program Selection" box, you can indicate how accurate of a match your submitted nucleotide sequence will make to a nucleotide sequence in your selected database. If you have no idea, you may want to start with "Highly similar sequences", perform the BLAST, and see what your results are. If you don't get any good matches, you may want to make the matching algorithm less rigorous. If you want to know more about different matching algorithms, there is a note on them at the bottom of this page. After you've chosen your matching algorithm, click BLAST. |
|
|
After you've clicked BLAST, you will probably have to wait a few minutes to get your results. The wait page looks like this: |
|
|
The results page is packed with data for you to use to quantify how good your BLAST match was. At the top of the page you will see a color-coded score alignment of your submitted sequence with all the sequences found in the database. The higher the score, the better. |
|
|
If you scroll down, you will see your top match. The hit tells you the species name of the species it aligned to, the name of the gene it aligned to (if applicable), the length of the gene, if the sequence was found from mRNA, the score of the match, the expect value (in microbiology slang, the e-value) of th ematch, the identities of the nucleotide match, the number of gaps, which strand the match occurred on, and then the actual alignment. This is a perfect match of my query sequence with reverse transcriptase from C. elegans: There are 1799/1799 identities and no gaps. To learn more about scores and e-values, please see the bottom of this web page. |
|
How to use BLAST Front Page
Genomics Page
Biology Home Page
Samantha's Home Page
Halorhabdus utahensis Genome Wiki