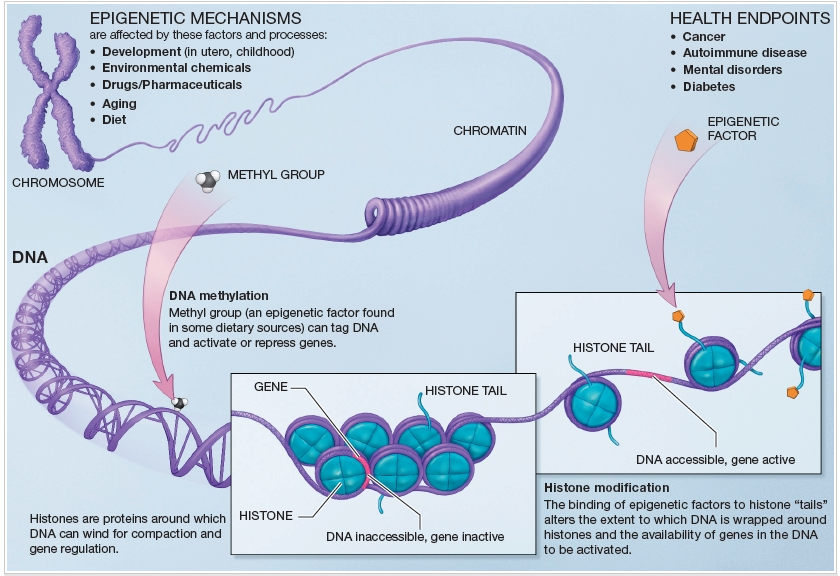
Epigenomics is the field that tries to answer the question of, “how the many cell types of the body maintain drastically different gene expression patterns while sharing the same DNA?”(Bethesda, 2010) Epigenomics is defined as, "the study of epigenetic modifications across the whole genome," (Bonneta, 2008). Epigenetic modifications can be made through DNA methylation and histone acetylation, which influence DNA coiling. These modifications help determine whether a gene is turned on or off in a cell. These changes can happen due to aging, diet, aging, chemicals, drugs and even during develpment. The genes may control anything from basic functions like cell division, to oncogenes (cancer-causing genes) and is connected to diseases such as cancer, diabetes, autoimmune diseases and much more (Fig 1.). By studying genomics scientists hope to understand which genes are turned on or off at certain times as well as in diseased tissues. With this knowledge, scientists might be able to get new insight into treating diseases and may begin to understand the complexities of the genome.
DNA methylation and histone acetylation are the two most important mechanisms of epigenomics. Histones are the proteins that DNA winds around to compact into chromosomes containing certain chemical tags (Bonetta, 2008). When an acetyl group is added to a histone it does not let the DNA coil as tightly. Methylation of DNA provides the opposite affect. When the enzyme DNA methyltransferase methylates DNA it causes the DNA to coil more tightly. Figure 1 gives a big picture view of how adding a methyl group causes the DNA to wind tightly so that the mRNA cannot be transcribed. It also shows that the addition of an epigenetic factor (like an acetyl group) can cause the DNA to unwrap allowing RNA polymerase to bind and make mRNA. These epigenetic modifications can have profound effects on expression of genes in different tissues and influence how tightly the chromosomes are wound.

Fig 1. Illustrates how epigenomic factors and DNA methylation can influence DNA coiling around histones and how this coiling influences how tightly the DNA is wound. (Courtesy of the NIH)
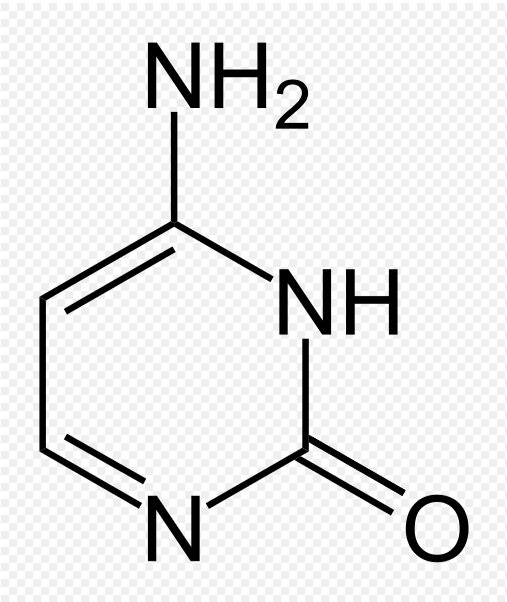
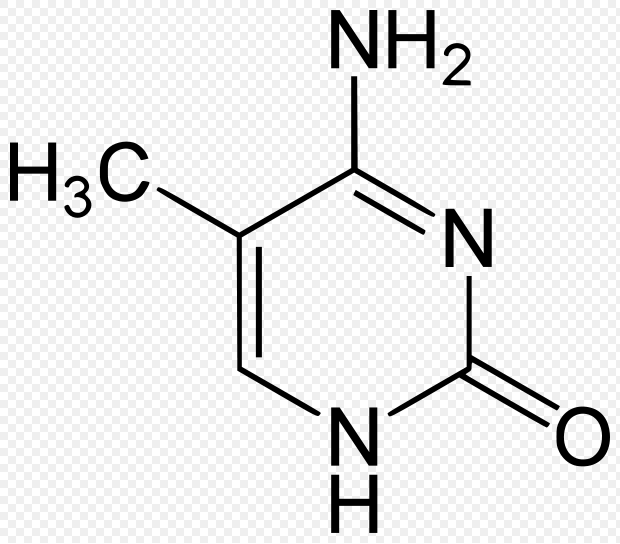
Fig 2. The molecule on the left is cytosine and the molecule on the right is 5-methylcytosine (methylated cytosine). Coutesy of Wiki Commons
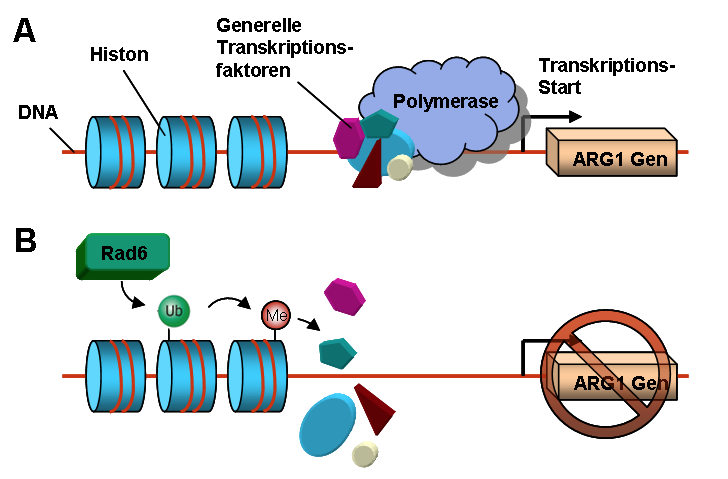
Fig 3. This figure demonstrates how adding a methyl group causes a change in DNA coiling that prevents the polymerase from binding, therefore preventing the gene (ARG1) from being transcribed. Image courtesy of Wiki Commons.
Scientists have developed many methods of studying epigenomics including microarrays, Genetic and Epigenomic Wide Association Studies (GWAS/EWAS), and methylated DNA Immunoprecipitation (MeDIP).
Microarrays are used to determine what genes are turned on and off in certain tissues at certain times. A microarray is composed of a well plate with DNA from different tissues, the same tissues at different time or from different people amplified in each well. Fluorescent probes are created from mRNA that can bind to the genes of interest. If the gene of interest is turned on in the tissue the probe will bind and let out light, if a probe does not bind the gene is turned off. They can probe multiple spots at once using different color probes. This allows many tissues and many genes to be tested quickly.

Fig 4. An example of a microarray courtesy of Wiki Commons
GWAS looks at certain loci over large populations to make correlations between disease and sequencing patterns. This idea has also been applied to the epigenome in Epigenomic Wide Association Studies (EWAS), however they have had limited success.
Methylated DNA Immunoprecipitation (MeDIP) takes microarrays one-step further. You make antibodies to 5-methylcytosine (methylated cytosine). After cutting genomic DNA with some sort of restriction enzyme you can mix the antibodies in so that they can bind to methylated segments of DNA. These can be purified and amplified, then labeled fluorescently. The fluorescent DNA can be placed on a microarray, when the probes from certain parts of the genome bind a signal is produced. Using this signal, the methylation can be located over the entire genome.
Bisulfite treatment is another common method. This technique involves changing unmethylated cytosine into uracil then amplifying only the DNA sequences that have 5-methylcytosines. This treated DNA can be used as a probe to determine if a section of DNA is methylated or not in the genome.
Twin studies are also very useful when it comes to epigenomic research. A set of monozygotic (identical) twins can have their DNA flouresently marked for methylation. In a study by Fraga et al. methylation changes were studied in two sets of monozygotic twins, which means the individuals started out with the same genetic information. One set was 50 years old and the other was 3 years old. They found that the 50 year old twins had more variation in methylation than the 3 year old twins, which suggests that environmental factors and aging can cause methylation changes that are not pre determined. (Fraga et al. 2005). A figure of the tagged chromosomes can be viewed here.
Epigenomics and disease have been related in many different studies. There is a lot of promise for advances in medicine using Epigenomics. The goal of this research is to create personalized medicine to help improve effectiveness of medicine. For example tumor suppressors and oncogenes have been linked to changes in methylation. MeDIP is often used to find these specific sites of methylation. If epigenetics were understood better, medications could be produced cure cancer and many other diseases.
Environmental factors also affect the epigenome. A study by Dolinoy et al demonstrated how the environment can influence the genome. They showed that in utero exposure to bisphenol A (BPA), which is used in plastics, is associated with higher body weight, increased breast and prostate cancer, as well as disrupted reproduction. Exposure results in increased DNA methylation, which alters the offspring’s epigenome for its life. The study showed that epigenetic patterning during early stem cell development is sensitive to BPA. (Dolinoy et al., 2007).
Epigenome project: The Hunan Epigenome’s website states that, “The Human Epigenome Project (HEP) aims to identify, catalogue and interpret genome-wide DNA methylation patterns of all human genes in all major tissues.” They are looking to identify Methylation variable positions (MVPs) which are methylation markers that vary at different loci from person to person. They are very similar to SNPs which are loci that have a different numbers of nucleotide repeats depending on the individual. They hope to identify the MVPs so that these markers can be used to diagnose disease and help create more effective treatments for the future.
There is also another project run by the NIH called The Roadmap Epigenomecs Project. Thire website states their goal as, “producing a public resource of human epigenomic data to catalyze basic biology and disease-oriented research.” They hope to create reference epigenomes and hopes to make this raw data more accessible. This site is definitely worth taking a look at. It has a plethora of data to brows as well as some genomic tools to sort your own data. The idea of sharing data and tools is a movement that will allow more knowledge to be generated more quickly and more cost effective. This website can be viewed here.
Bethesda. 2010. Epigenomics Scientific Background. National Center for Biotechnology. http://www.ncbi.nlm.nih.gov/books/NBK45788/. January 2014.
Bonetta, L. 2008. Epigenomics: the new tool in studying complex diseases. Nature Education (1) 1:178
Dolinoy, D. 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. PNAS. 104 http://www.pnas.org/content/104/32/13056.full. January 2014.
Fraga, M., Ballestar, E., Paz, M., Ropero, S., Setien, F., Ballestar, M., Heine-Suner, D., Cigudosa, J., Urioste, M., Benitez, J., Boix-Chornet, M., Sanchez-Aguilera, A., Ling, C., Carlsson, E., Poulsen, P., Vaag, A., Stephen, Z., Spector, T., Wu, Y., Plass, C., and Esteller, M. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. The National Academy of Sciences 102:10604-10609.
National Human Genome Research Institute. 2014. Genome-Wide association Studies. National Institutes of Health. http://www.genome.gov/20019523. January 2014.
NIH. 2014. Roadmap epigenomics project. http://www.roadmapepigenomics.org/. January 2014.
Pospisilik, J. 2013. Bridging epigenomics and complex disease: the basics. Cellular and Molecular Life Sciences. 70:1609-1621.
Gracie's Page
Genomics Page
Biology Home Page
Email Questions or Comments: grgordon@davidson.edu.
© Copyright 2014 Department of Biology, Davidson College, Davidson, NC 28035